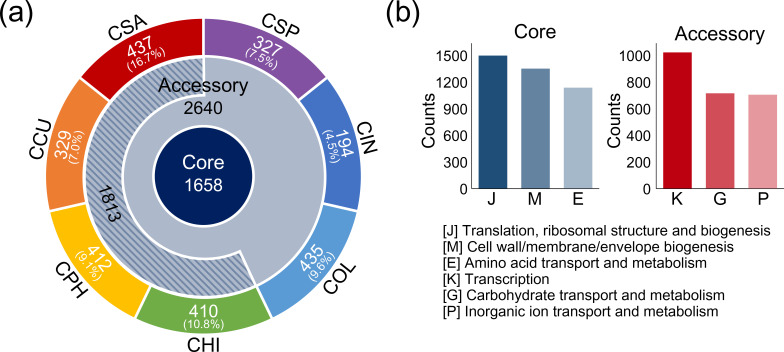
Fig. 3.
The pan-genome of seven Chryseobacterium species. (a) The pan-genome consisting of 29 340 genes was determined using PGAP and used to build a Venn diagram. The number in the centre ring is the number of gene clusters shared by all species, whereas gene clusters common among several species are shown in the peripheral ring (accessory component). The numbers in the outer rings correspond to the number of non-orthologous gene clusters specific to each species. The numbers in parentheses indicate the ratio of unique genes to the respective total genes. The number in the striped area represents the number of gene clusters shared by halotolerant species in the accessory genome. Acronyms used for Chryseobacterium species: CCU, C. cucumeris GSE06; CHI, C. hispalense DSM 25574; CIN, C. indologenes PgBE177; COL, C. oleae DSM 25575; CPH, C. phosphatilyticum ISE14; CSA, C. salivictor NBC122; and CSP, Chryseobacterium sp. T16E-39. (b) The COG database was used for functional analysis of the pan-genome. Annotated genes were classified into 24 functional categories. The top three categories in the core and accessory genomes are represented as bar graphs. The x-axis indicates each COG category, and the y-axis represents the number of genes.