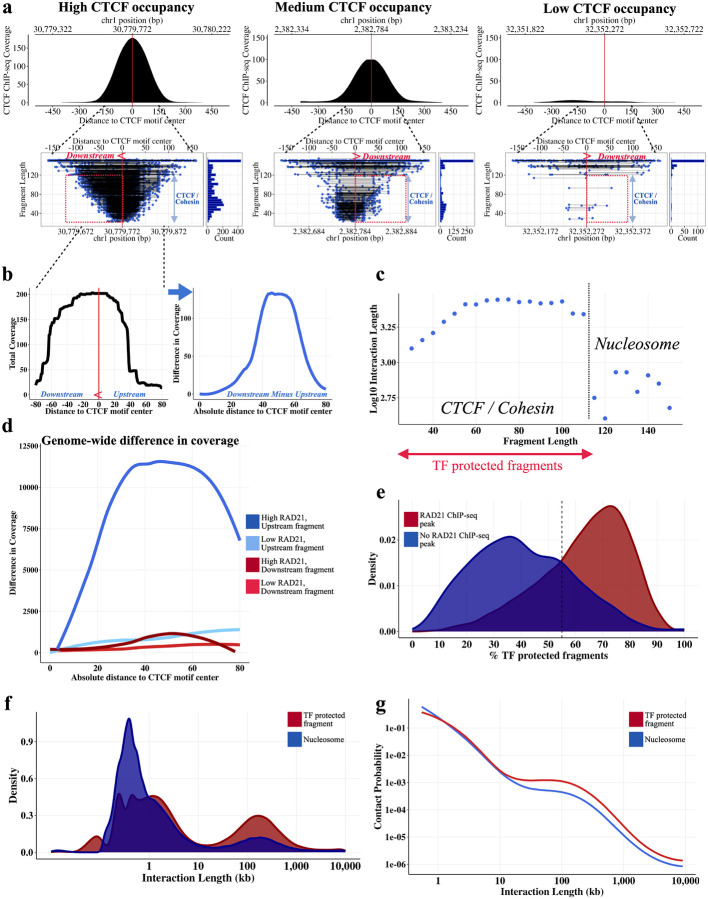
Fig. 4.
Cohesin and CTCF-protected fragments identified in CTCF MNase HiChIP. a High, medium, and low CTCF occupied motifs. Cohesin footprint is observed downstream of the CBS for high and medium CTCF occupancy motifs. For each occupancy level, CTCF ChIP-seq (top) and all fragments overlapping the CTCF motif (bottom left) are depicted, along with the corresponding fragment length histogram (bottom right). b Locus-specific high CTCF occupancy figure from (a) as a coverage plot (left figure), difference in coverage between downstream and upstream coverage (right figure). c Plotting median log10 interaction length as a function of fragment length suggests presence of nucleosome vs TF-protected fragments. Only left fragments overlapping CTCF (+) motifs with start and end at least 15 bp from the CTCF motif were included in this graph to remove confounding by MNase cut site. Using this figure, we are approximating CTCF +/− cohesin-protected fragments as those with fragment length < 115, start and end at least 15 bp from the motif center. d Difference in coverage (downstream - upstream) across all CBS shows an increase in coverage downstream of the CTCF motif for upstream fragments underlying CBS with a strong adjacent RAD21 ChIP-seq peak. e CTCF motifs that have a nearby RAD21 ChIP-seq peak (within 50 bp) have a larger proportion of TF-protected fragments. f TF-protected fragments have a noticeably larger bump in density of long range interactions compared to nucleosome-protected fragments. Fragments were first filtered to those with start and end at least 15 bp from the motif. TF-protected fragments were then defined as fragments with length < 115 bp while nucleosome-protected fragments are fragments with length at least 115 bp. g P(S) curve for fragments depicted in (f).