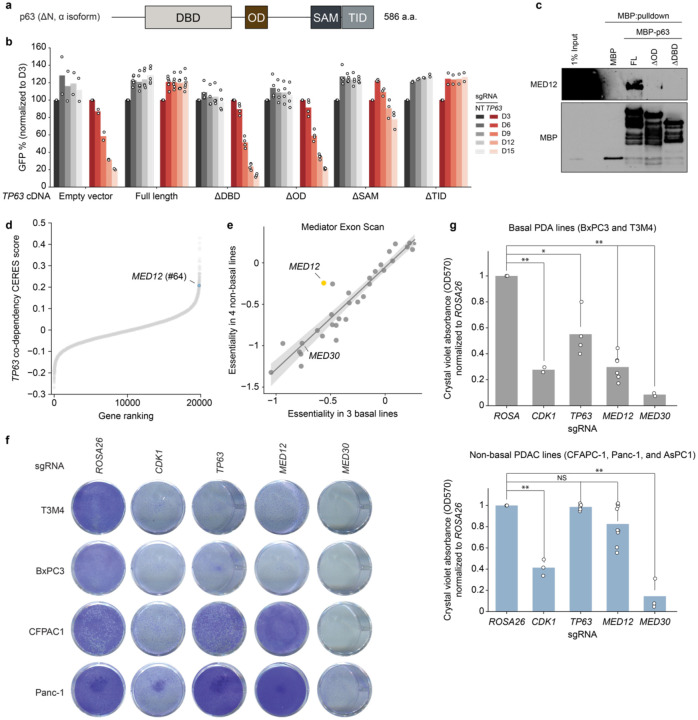
Fig. 4. MED12 is a lineage-biased genetic vulnerability of basal-like PDAC.
a, Diagram of p63 (N-terminus ΔN, C-terminus α isoform) domain architecture. b, Gene complementation competition-based proliferation assay of different overexpressed p63 truncation mutants upon endogenous TP63 CRISPRi-induced knockdown. c, MBP pulldown assay using purified MBP, full length MBP-ΔNp63 (FL), or MBP fusion p63 mutants lacking the oligomerization domain (ΔOD) or DNA-binding domain (ΔDBD). Purified protein was incubated with MED12-expressing Sf9 lysates overnight, followed by pulldown and western blot. d, Pearson r of >17.000 DepMap CERES gene codependencies with TP63 across ~1000 cell lines. e, Scatterplot of mean essentiality scores of all sgRNAs targeting each of 33 genes in the Mediator complex in basal (T3M4, BxPC3, and PK-1) and non-basal (SUIT2, AsPC-1, KLM1, and MiaPaCa-2) cell lines. Linear regression and 95% confidence interval are drawn. f, Crystal violet staining of basal (T3M4, BxPC3) or non-basal (CFPAC1, Panc-1) human PDA lines grown for 10 days after knockout of negative control (ROSA26), lethal control (CDK1), TP63, MED12, or panessential Mediator gene MED30. b, Quantification of re-solubilized crystal violet stain of basal (T3M4, BxPC3; top) or non-basal (CFPAC1, Panc-1, AsPC-1; bottom) grown for 10 days, as quantified by OD570. Two-sided t-test p-values: * < 0.05; ** <0.01; NS not significant.