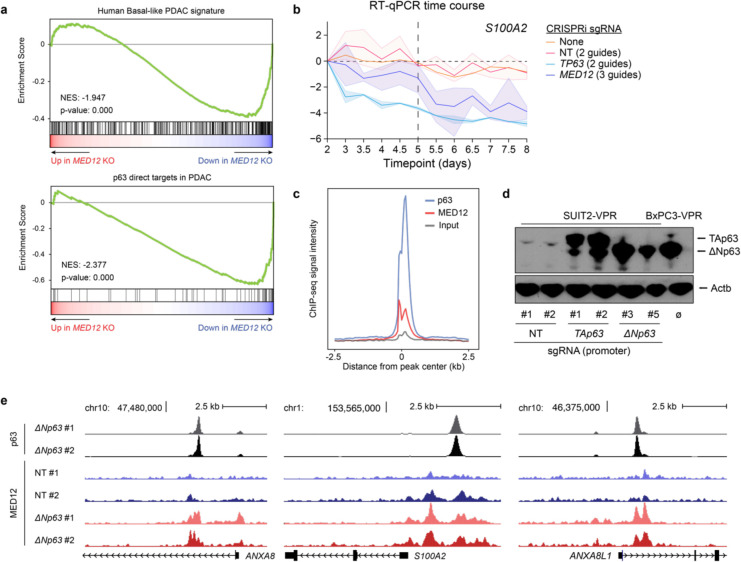
Extended Fig. 2. MED12 and p63 co-regulate the basal gene expression program.
a, Representative GSEA plots of KLM1 MED12 knockout using gene signatures derived from human basal PDAC tumors (Chan-Seng-Yue et al. 2020) and direct p63 gene targets in PDAC (Somerville et al. 2018). The plots were generated and the normalized enrichment scores (NES) and statistics were calculated using DESeq2-derived log2FC values of MED12 vs ROSA26 KO in GSEApy. Three biological replicates were used for each sample. Complete GSEA analysis for all the sgRNA tested can be found in Supplementary Table 2. b, Time-course RT-qPCR of S100A2 after lentiviral infection with CRISPRi sgRNA targeting TP63 (2 sgRNA), MED12 (3 sgRNA), non-targeting sgRNAs (2 sgRNA), or uninfected control T3M4 cells. −ΔΔCt values are plotted as the average of each sgRNA normalized to the average of housekeeping genes ACTB and B2M. For each gene perturbation, the average −ΔΔCt value is shown in a solid line, and the 95% confidence intervals are shown as translucid intervals. MED12 knockdown inflection point (~ day 5) is marked by a vertical black dashed line. c, Metaplot of genomic occupancy of p63 and MED12 centered around p63 peaks in T3M4 cells. d, Western blot of SUIT2 CRISPR-activated TP63 (isoform-specific) cells. BxPC3, which endogenously expresses the ΔN isoform of p63, is shown as a positive control in the rightmost lane. e, Genomic tracks of p63 and MED12 occupancy at direct p63 targets ANXA8, S100A2, and ANXA8L1 in SUIT2-VPR lines infected with non-targeting (NT) or TP63-targeting sgRNAs.