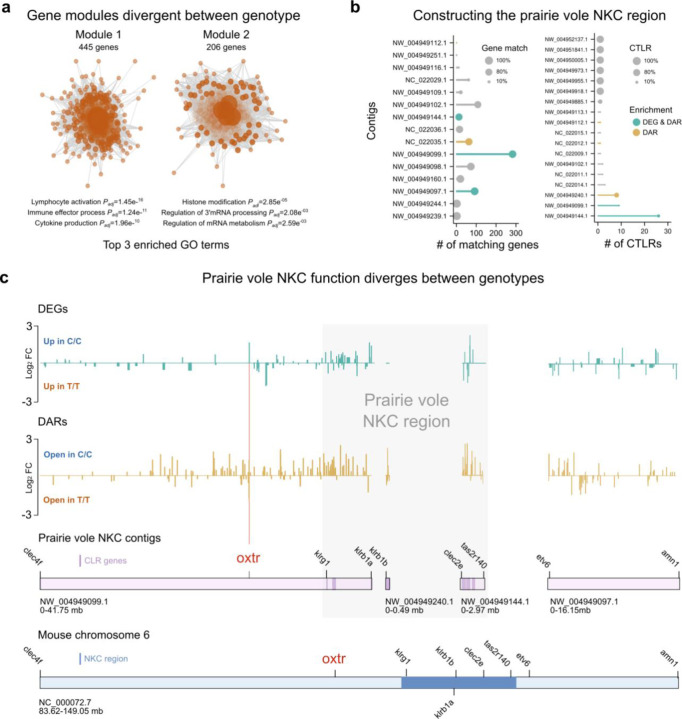
Figure 2. NKC function is divergent between genotypes.
a) Schematic illustrating two gene modules that are significantly different between genotypes, with top 3 GO terms indicated. b) (Left) Plot shows the number of annotated genes that a given prairie vole scaffold shares with the mouse Oxtr chromosome. scaffolds with no matching genes are not included. Dot size represents the percentage of matching genes. (Right) Plot depicts number of CTLR genes on a given prairie vole scaffold. Scaffolds without CTLRs are not included. Dot size indicates the percentage of genes that are CTLRs. c) Upper two plots depict the fold changes of DEGs (green traces) and DARs (gold traces) across NKC scaffolds. Lower plots illustrate how NKC vole scaffolds map to the mouse Oxtr chromosome. Scaffolds are on scale, but scales of vole and mouse scaffolds differ.