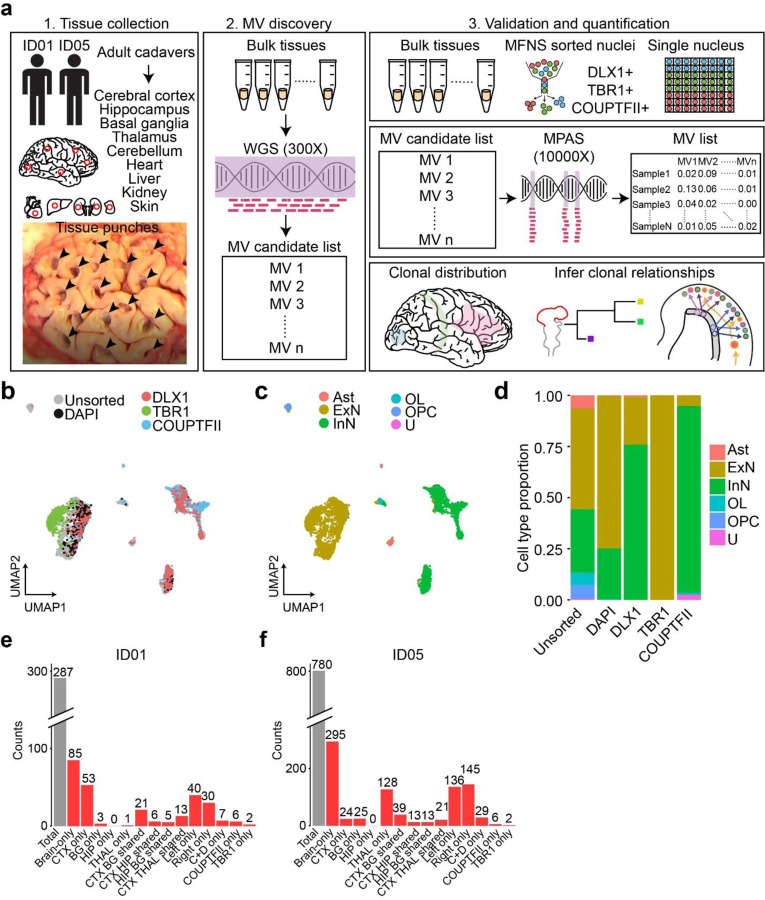
Figure 1. Comprehensive cMVBA identifies cell-type-resolved and region-specific MVs.
(a) cMVBA workflow overview consists of three phases: 1. Tissue collection: Cadaveric organs accessed for tissue punches from organs listed (red circles and black arrows: punch locations in organs and frontal lobe) for MV detection. 2. A subset of the bulk tissue punches undergo 300x WGS followed by best-practice MV calling pipelines to generate a list of MV candidates. 3. DNA from each punch bulk tissue, methanol fixed nuclear sorted (MFNS) samples, or individual nuclei are subjected to validation and quantification of MV candidates via massive parallel amplicon sequencing (MPAS). AFs of the validated MVs from MPAS are used to determine clonal dynamics of different neuronal cell types and reconstruct features of brain development. (b) UMAP plot from snRNA-seq with MFNS sorted or unsorted cortical nuclear pools (n = 3322). Sorted nuclear groups labeled with distinct colors. (c) Cortical cell types based on marker expression and differentially expressed genes in each cluster. (d) Cell type proportion within each sorted cortical nuclear population. (e–f) Number of MVs categorized by location detected in each donor ID01 or ID05 (Supplementary Data 4). ‘Brain-only’ MVs (i.e., detected only in brain tissue, but not other organs) including subtypes in red. ‘C+D only’: Brain-only MVs exclusively detected in both COUPTFII+ and DLX1+ populations but not the other cell types. Ast, Astrocyte; ExN, Excitatory neurons; InN, inhibitory neurons; OL, oligodendrocytes; OPC, oligodendrocyte precursor cells; U, undefined.