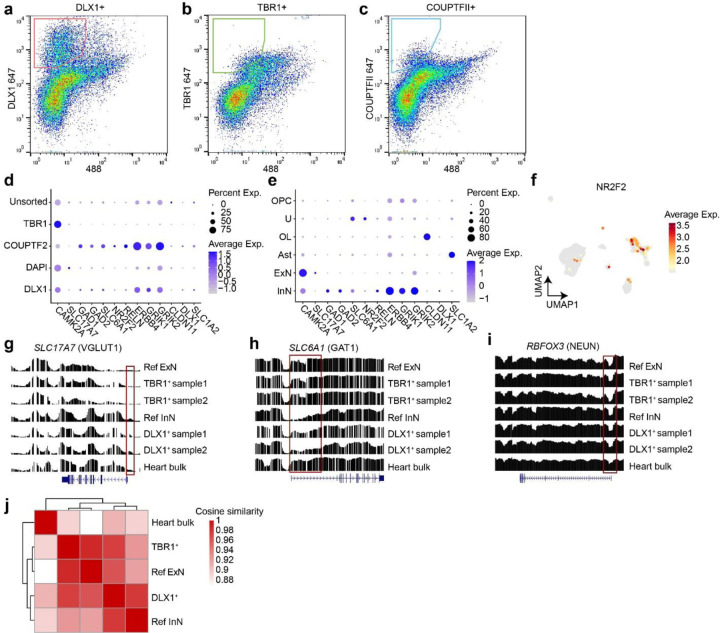
Extended Data Fig. 2. Bisulfite sequencing in sorted TBR1+ and DLX1+ nuclear pools correlate with excitatory and inhibitory neuron methylome signatures.
(a–c) MFNS gating strategy on 30,000 single brain nuclei using DLX1, TBR1, and COUPTFII antibodies. (d) snRNA-seq of post-MFNS nuclei confirming enrichment of targeted nuclear types. (e) Marker expression in assigned nuclear types correlating with targeted nuclear types. (f) UMAP plot NR2F2 expression pattern (encoding COUPTFII) highlighting a subpopulation of inhibitory neurons (compare with Fig. 1c). (g–i) Reference excitatory and inhibitory neuronal methylome signatures (aggregated from an available public single-nuclei methylome dataset31) compared to methylome signatures of sorted nuclei and a bulk heart sample. Normalized relative methylation levels (y-axes) and genomic positions (x-axes) of genes listed at top. (g) Methylome signature of SLC17A7 encoding VGLUT1, an excitatory neuronal marker in the brain, showing reduced methylation (i.e. representing activation) across the gene body and especially near the transcription start site (TSS, red box) in TBR1+ excitatory neuron samples. (h) Methylome signature of SLC6A1 encoding VGAT1, an inhibitory neuronal marker in the brain, showing reduced methylation across the gene body and especially near the TSS (red box). (i) Methylome signature of RBFOX3 encoding NEUN, a mature neuronal marker in the brain, showing reduced methylation at the TSS in neurons compared with bulk heart. Ref ExN, reference excitatory neurons; Ref InN, reference inhibitory neurons. (j) Heatmap and dendrograms based on cosine similarities of global methylation patterns between groups. Two different TBR1+ or DLX1+ samples were aggregated. The TBR1+ nuclear pool was clustered with Ref ExN while the DLX1+ clustered near the pool with Ref InN. The control heart bulk sample was distant from either group.