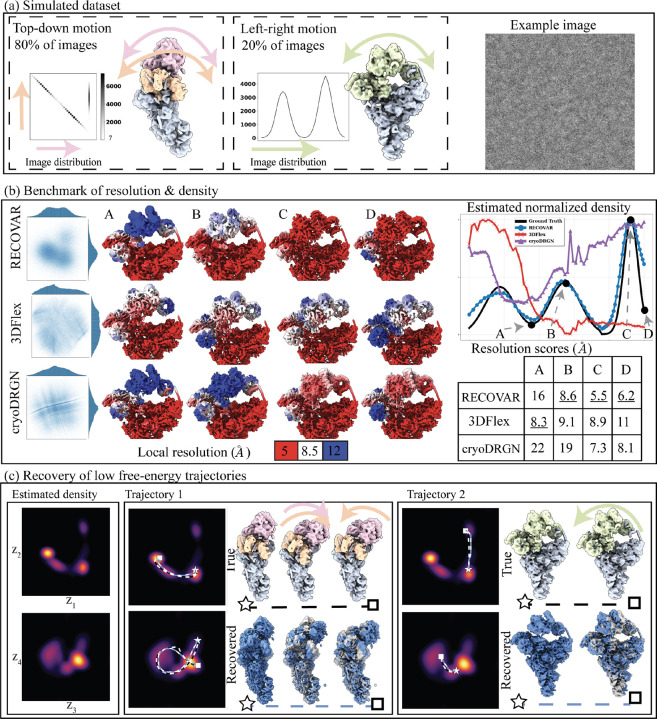
Figure 2:
Validation of RECOVAR on a synthetic dataset. (a) Simulated dataset of the precatalytic spliceosome with three bodies moving independently. A landscape of motions consisting of 80% images where the orange and pink bodies move independently in top-down motions, including a high-density trajectory where both bodies first move down together, and the orange body then locks back into place. A second motion, consisting of 20% of images, shows the two bodies’ left-right motion. (b) Benchmarking of RECOVAR against two deep-learning methods 3DFlex and cryoDRGN. The embedding along two PCs of the embedding is shown for each method, and 4 reconstructed states are plotted in their local resolution. The 90% percentile of the local resolution for each state is shown in the bottom right. The estimated density along the high-density top-down motion of each method is shown in the top right. (c) (Left) The estimated 4-dimensional density of the RECOVAR embedding is displayed over pairs of dimensions by integrating the remaining dimensions. (Middle and right) The two ground truth (in black) and two recovered (in blue) trajectories are shown in volume and latent space. The preceding state along the trajectory is overlaid in gray for comparison. The star and the square represent the endpoints.