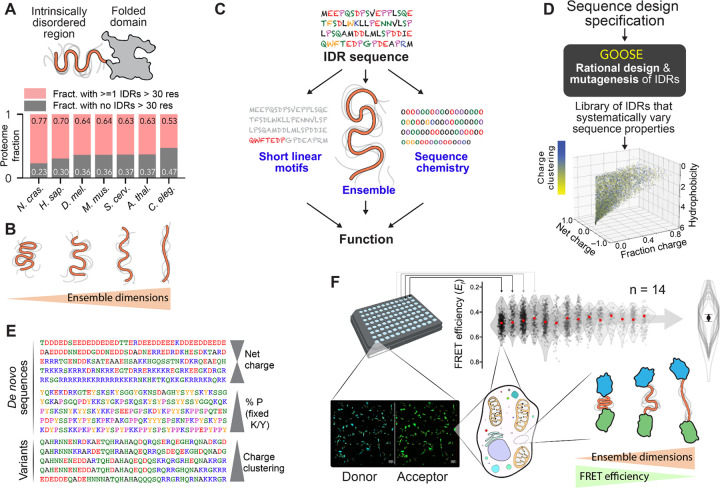
Figure 1. IDR ensemble properties can be important determinants of function.
(A) IDRs are protein regions that lack a stable 3D structure and are ubiquitous across eukaryotic proteomes. (B) While IDRs lack a fixed 3D structure, their conformational behavior can be quantified in terms of ensemble dimensions. (C) Emerging work suggests that IDR function depends on an interplay between three key features: sequence chemistry, ensemble properties, and the presence of short linear binding motifs (SLiMs). (D) GOOSE is a computational tool for the rational design or mutagenesis of IDRs. In the scatter plot, each point represents a sequence designed by GOOSE to match the constraints defined by the axes and in the color bar. (E) GOOSE enables specific sequence parameters to be titrated while others are held fixed. (F) An in-cell fluorescence reporter assay combined with a library of rationally designed sequences enables us to determine how IDR sequence properties dictate ensemble properties in the cellular environment (bottom right). Experiments are performed using a 96-well imaging plate (top left), and each well with 60 or more cells is analyzed to calculate FRET efficiency (Ef) based on donor and acceptor fluorescence (bottom left). Across multiple independent measurements (14 in this case), average EF values are calculated to provide a statistically rigorous and highly reproducible assessment of sequence-specific FRET efficiency (top right). Only the summary violin plot overlays shown here are reported in all other figures, with the average median shown as a circle and the SD of medians as error bars.