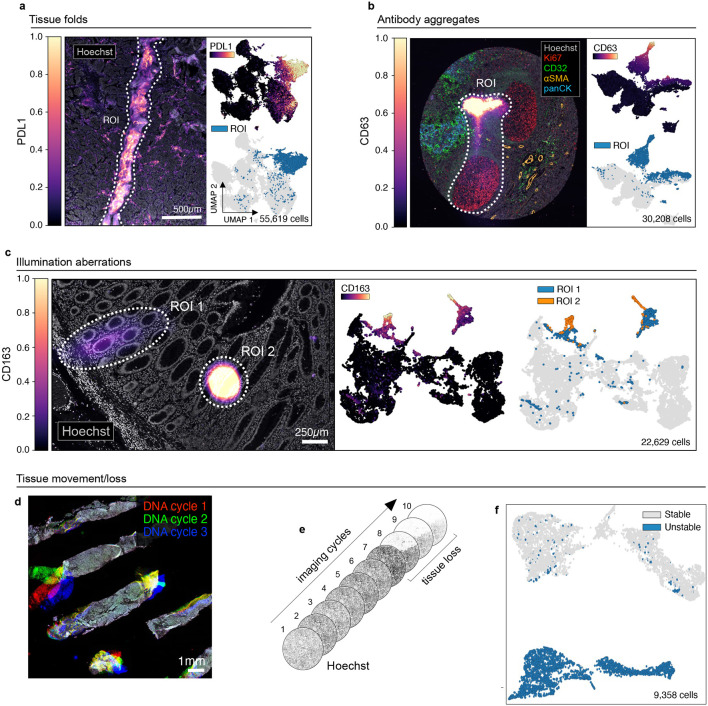
Fig. 1 ∣. Recurring artifacts in whole slide immunofluorescence images of tissue and the effects on tissue-derived single-cell data.
a, Left: Tissue folding ROI (dashed white outlines) in TOPACIO sample 110 as viewed in the PD-L1 (colormap) and Hoechst (gray) channels. Right: UMAP embeddings showing cells in the image colored by PD-L1 intensity (top) and whether they fall into the ROI shown at left (bottom). b, Left: Antibody aggregate in the CD63 (colormap) channel of EMIT TMA core 68 (human tonsil). Hoechst (gray), Ki67 (red), CD32 (green), αSMA (orange), and pan-CK (blue) are shown for context. Right: UMAP embedding showing cells from the image colored by CD63 intensity (top) and whether they fall within the ROI shown at left (bottom). c, Illumination aberrations in the CD163 (colormap) channel of the CRC dataset. Left: Image showing two bright spots (dashed white ROI outlines); Hoechst (gray) shown for reference. Right: UMAP embeddings showing 22-channel data from cells in the ROIs colored by CD163 intensity (left UMAP plot) and whether they are within one of the two ROIs shown in the corresponding image (right UMAP plot). d, Tissue detachment in TOPACIO sample 80 as demonstrated by superimposing Hoechst signals from three different CyCIF imaging cycles: 1 (red), 2 (green), 3 (blue). e, Progressive tissue loss in EMIT TMA core 1 (normal kidney cortex) across 10 imaging cycles as observed in the Hoechst channel (gray) where overt tissue loss is seen by cycle 8. f, UMAP embedding of cells from EMIT TMA core 1 (normal kidney cortex) colored by whether cells remained stable (gray data points) or became detached (blue data points) over the course of imaging demonstrating that unstable cells form discrete clusters in feature space.