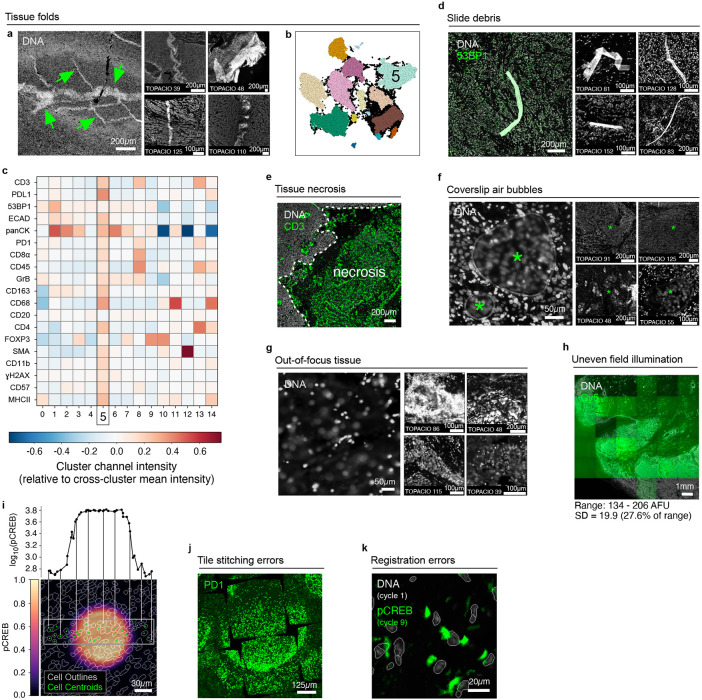
Extended Data Fig. 2 ∣. Recurring artifacts in whole slide immunofluorescence images of tissue and the effects on tissue-derived single-cell data.
a, Tissue folding (green arrows) in normal human tonsil tissue imaged by mIHC as seen in the Hoechst channel (gray). Four additional examples of tissue folding in TOPACIO samples are shown at right. b, UMAP embedding shown in Fig. 1a colored by HDBSCAN clusters demonstrating the formation of a discrete cluster (cluster 5) caused by the tissue fold. c, Heatmap showing the difference between the channel signal intensities of individual clusters shown in Fig. 1a and Extended Data Fig. 2b and the average channel signal intensity across all clusters demonstrating that cluster 5 (corresponding to the tissue fold) exhibits greater-than-average intensities in all 20 channels in the image. d, Autofluorescent fiber seen in the 53BP1 (green) and Hoechst (gray) channels of TOPACIO sample 128. Four additional examples of slide debris in TOPACIO samples are shown at right. e, Necrosis in a region of tissue from TOPACIO sample 39 as seen in the CD3 (green) channel. f, Coverslip air bubbles (green asterisks) in TOPACIO sample 48 as seen in the Hoechst channel (gray). Four additional examples of coverslip air bubbles in TOPACIO samples are shown at right. g, Out-of-focus region of tissue in TOPACIO sample 55 as seen in the Hoechst channel (gray). Four additional examples of out-of-focus tissue in TOPACIO samples are shown at right. h, Uneven tile illumination in the HNSCC CODEX dataset as seen in the Cy5 channel. Hoechst (gray) shown to highlight tissue location. The standard deviation among per-tile median signal intensities was 19.93 arbitrary fluorescence units (AFU), 27.6% of the range (134-206 AFU). i, Illumination aberration in the pCREB (colormap) channel of EMIT TMA core 95 (dedifferentiated liposarcoma) with nuclear segmentation outlines (translucent contours) shown for reference. The line plot above shows the pCREB signal of cells highlighted by green scatter points in the image below. j, Tile stitching errors in the mIHC dataset of normal human tonsil as seen in the PD1 (green) channel. k, Cross-cycle image registration error in EMIT TMA core 64 (leiomyosarcoma) as demonstrated by superimposing the Hoechst (gray) signal from imaging cycle 1 and the pCREB (green) signal from imaging cycle 9.