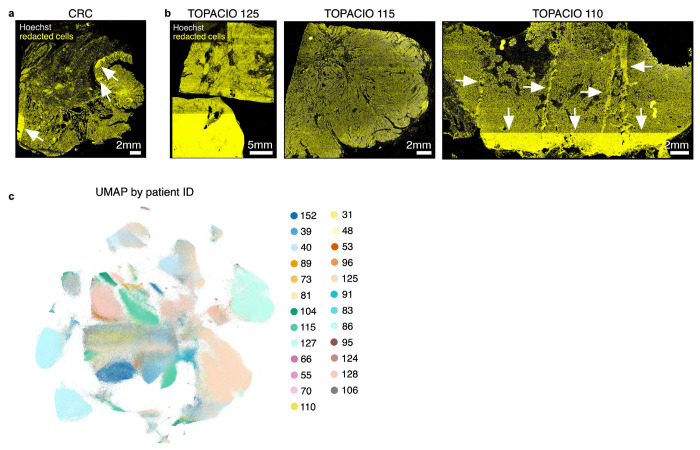
Extended data Fig. 6 |. Location of cells redacted by CyLinter in Dataset 2 (CRC) and Dataset 1 (TOPACIO) and Post-QC TOPACIO UMAP embedding colored by patient ID.
a, Cells redacted by CyLinter from the Dataset 2 (CRC) demonstrating no discernable bias in the removal of cells from the image with the exception of those areas highlighted by the white arrows which were affected by focal artefacts and removed using CyLinter’s selectROIs module. b, Cells redacted by CyLinter from three arbitrary specimens from Dataset 1 (TOPACIO) demonstrating no discernable bias in the removal of cells from the images with the exception of those areas highlighted by the white arrows which were affected by focal artefacts and removed using CyLinter’s selectROIs module. c, UMAP embedding of post-QC TOPACIO data shown in (Fig. 6b) colored by specimen ID demonstrating patient-specific clustering in tumor cell populations, but not immune and stromal populations (refer to Fig. 6b,d,e–i and Online Supplementary Fig. 8 for cluster phenotype identities).