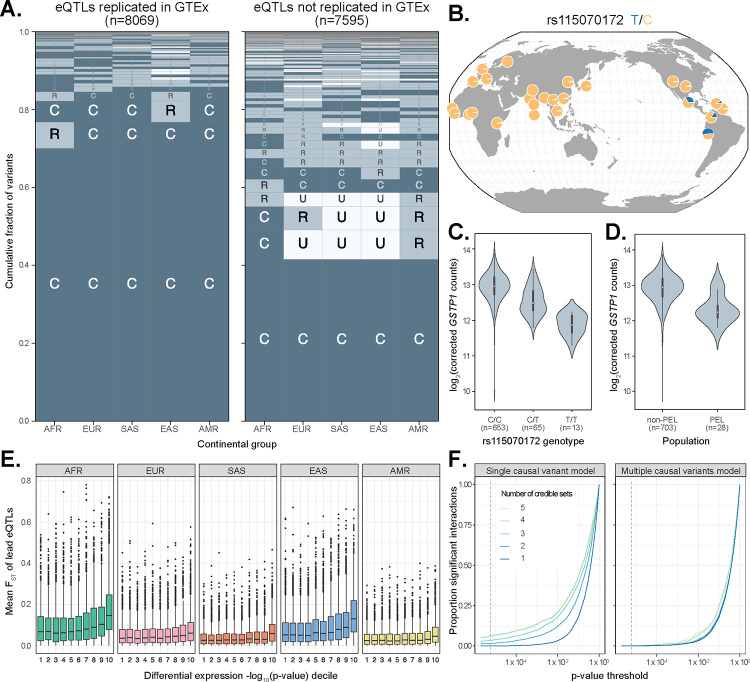
Figure 5. Population-specific genetic effects on gene expression.
(A) Visualization of the joint distribution of allele frequencies of MAGE fine-mapped lead eQTLs across continental groups from the 1000 Genomes Project11, stratifying on replication status in GTEx15. (B) Allele frequency of a lead eQTL of GSTP1 (rs115070172) across populations from the 1000 Genomes Project. The minor (T) allele achieves high frequency (AF = 0.63) in the Peruvian (PEL) population but is at moderate or low frequencies in other global populations. (C) Expression of GSTP1, stratified by genotype of rs115070172. (D) Expression of GSTP1, stratified by population label (PEL versus non-PEL). (E) Frequency differentiation (measured as mean FST between the focal continental group and all other groups) of lead eQTLs, stratifying by differential expression decile (contrasting expression in the focal continental group versus all other groups) of respective eGenes, where the 10th decile represents the strongest evidence of differential expression based on p-value. Across all continental groups, evidence of differential expression is positively associated with levels of frequency differentiation of lead eQTLs. (F) Number of significant genotype-by-continental group interactions at varying p-value thresholds (dashed line denotes Bonferroni threshold) for a model that considers a single causal variant at a time (left panel) versus a model that jointly considers multiple potential causal variants (right panel). Results are stratified by the number of credible sets (from one to five), demonstrating that failure to consider multiple causal variants may erroneously manifest as ancestry-specific heterogeneity of genetic effects on expression.