Figure 3. Dominance variants localize to putative CXCL12 regulatory regions.
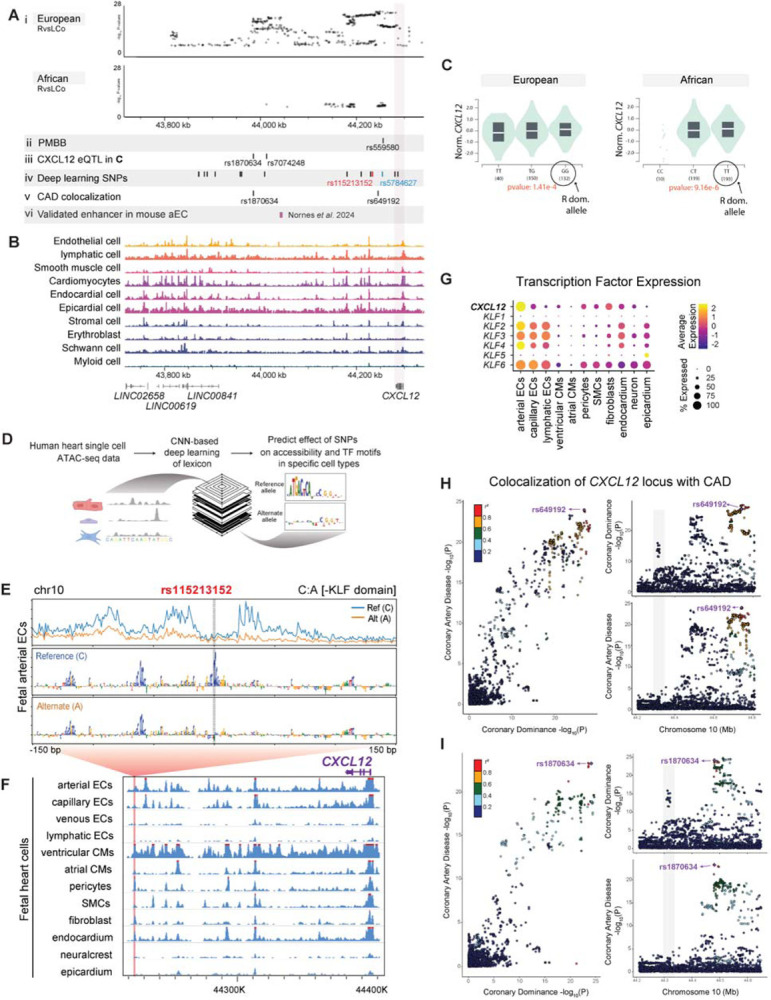
(A) SNPs reaching genome wide significance in the indicated MVP populations aligned with variants from additional analyses: Penn Medicine Biobank (PMBB) replication study, Genotype-Tissue Expression (GTEx) eQTLs, deep learning-based predictions, and coronary artery disease (CAD) colocalization. An in vivo validated Cxcl12 enhancer is also shown. (B) Location of accessible chromatin in human fetal heart cell types in relation to dominance SNPs in A. (C) Violin plots of two lead SNP eQTLs revealed that a higher prevalence of right dominance is associated with increased CXCL12 in GTEx data. Left is rs1870634; right is rs7074248. Testis expression levels by genotype are shown, but directionality is the same in arterial and coronary tissues (see Supplementary Figure 5A, B). (D) Schematic of convolutional neural network (CNN) deep learning models used to predict the effect of alternate alleles. (E) Predicted per-base accessibility for reference and alternate alleles of rs115213152 suggest a disrupted KLF motif. (F) Sequencing tracks of chromatin accessibility near CXCL12. Red highlights rs115213152 region where open chromatin coincides with CXCL12 promoter accessibility in select cardiac cell types, including coronary artery cells. (G) ScRNAseq from fetal hearts revealed co-expression of CXCL12 and KLFs, supporting a potential role for these transcription factors in the activity predicted in E. (H and I) LocusCompare ouput displaying colocalization plot (left), and LocusZoom plots for each trait (right). The SNP with the highest H4 posterior probability using COLOC is labelled in purple with additional variants in linkage disequilibrium with labelled SNP colored according to R2 value. Top panel (H) is unmodified analysis and bottom panel (I) shows result when the first colocalization signal at rs649192 (44650000–44850000) is masked.
