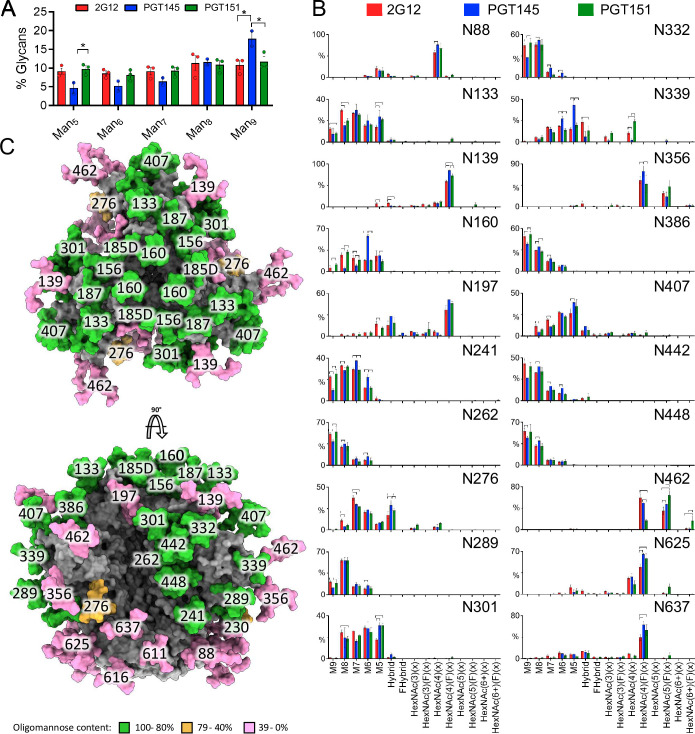
Fig 8. Global and site-specific fine glycan-type determination.
A. HILIC-UPLC analysis of Endo H-released and fluorescently labeled N-glycans from CZA97.012 SOSIP.664 trimer purified by 2G12, PGT145, or PGT151 and then by SEC. The bar graphs represent the abundance of each oligomannose-type glycan as listed on the category axis. The percentage on the y axis is proportion of each oligomannose-type glycan out of the total, i.e., PNGase F-released, glycan pool. B. CZA97.012 SOSIP.664 trimer was 2G12-, PGT145- or PGT151- and then SEC-purified. Site-specific glycan content was determined by LC-MS on intact glycopeptides. The proportion of each type of glycan is depicted on the y-axes (%), the glycan type on the horizontal category axis. Error bars show s.e.m. for 2 independent MS analyses. Significant differences within pairs of purified trimer preparations (p<0.05, t-test) are bracketed. C. Glycosylated model of a HIV-1 Env trimer (truncated C-terminally of residue 664), based on the site-specific glycan data for 2G12-purified material. The predominant glycan composition determined through the analysis displayed in panel B was modelled onto the trimer structure obtained by Cryo-EM of CZA97.012 SOSIP.664 in complex with Fab 3BNC117 and reported in this study (EMD-40088 and PDB 8gje). Where a site was not resolved by LC-MS, a representative glycan consisting of Man5GlcNAc2 was used instead. Individual glycan sites are coloured according to the abundance of oligomannose-type glycans at each site, as shown in the key.