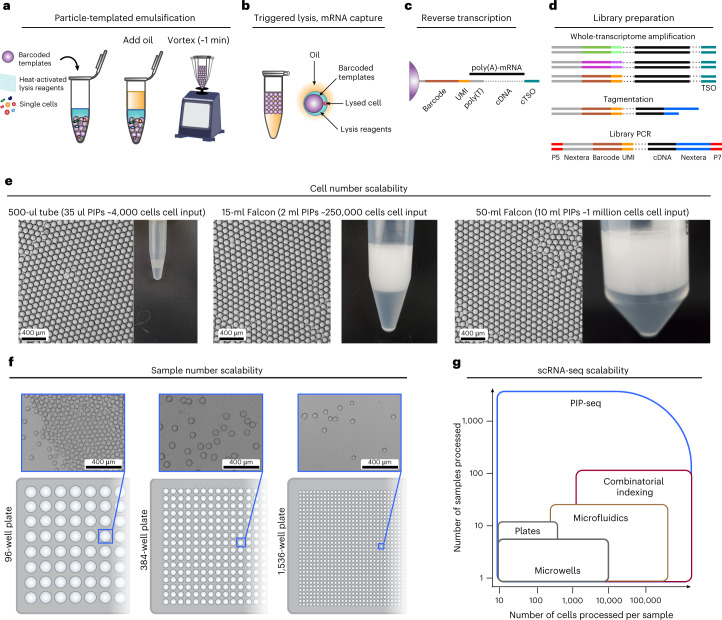
Fig. 1. Rapid and scalable templated emulsification for single-cell genomics.
a–d, PIP-seq enables the encapsulation, lysis and barcoding of single cells. a, Schematic of the emulsification process. Barcoded particle templates, cells and lysis reagents are combined with oil and vortexed to generate monodispersed droplets. b, Heat activation of PK results in lysis and release of mRNA that is captured on bead-bound barcoded poly(T) oligonucleotides. c, Oil removal is followed by bulk reverse transcription of mRNA into cDNA. cTSO is the complement of the template switch oligonucleotide. d, Barcoded whole-transcriptome-amplified cDNA is prepared for Illumina sequencing. e–g, Efficient single-bead, single-drop encapsulation at scale. e, Particle-templated emulsification in different-sized tubes (1.5 ml, 15 ml and 50 ml) produces monodispersed emulsions capable of barcoding orders of magnitude different cell numbers. f, PIP-seq is compatible with plate-based emulsification, including 96-, 384- and 1,536-well plate formats. Representative images are shown from experiments completed three times. g, The estimated ability of different technologies to easily scale with respect to cell and sample number.