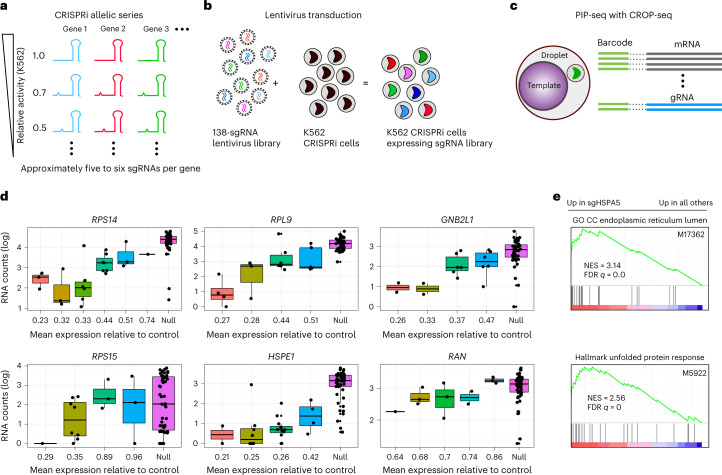
Fig. 4. Transcriptome and gRNA sequencing using PIP-seq.
a, Schematic of the CROP-seq sgRNA library designed with target mismatches to modulate the activity of essential genes. b, Lentiviral transduction of the CRISPRi library in K562 cells. c, Schematic of the capture and barcoding of polyadenylated mRNA and sgRNA using PIP-seq. RNA and sgRNA libraries are prepared separately and pooled for sequencing. d, Quantification of gene expression of sgRNAs within an allelic series. sgRNAs are ordered from high to low predicted knockdown efficiency35. Non-targeting sgRNAs are denoted as “Null”. Box plots indicate the median, with the lower and upper hinges corresponding to the 25th and 75th percentiles, respectively, and raw data points are displayed (with slight jitter). e, Preranked gene set enrichment analysis (GSEA) of scRNA-seq data comparing sgHSPA5-transduced cells to non-sgHSPA5-transduced cells shows enrichment in genes related to endoplasmic reticulum stress and unfolded protein response; GO CC, Gene Ontology cellular component; NES, normalized enrichment score; FDR, false discovery rate.