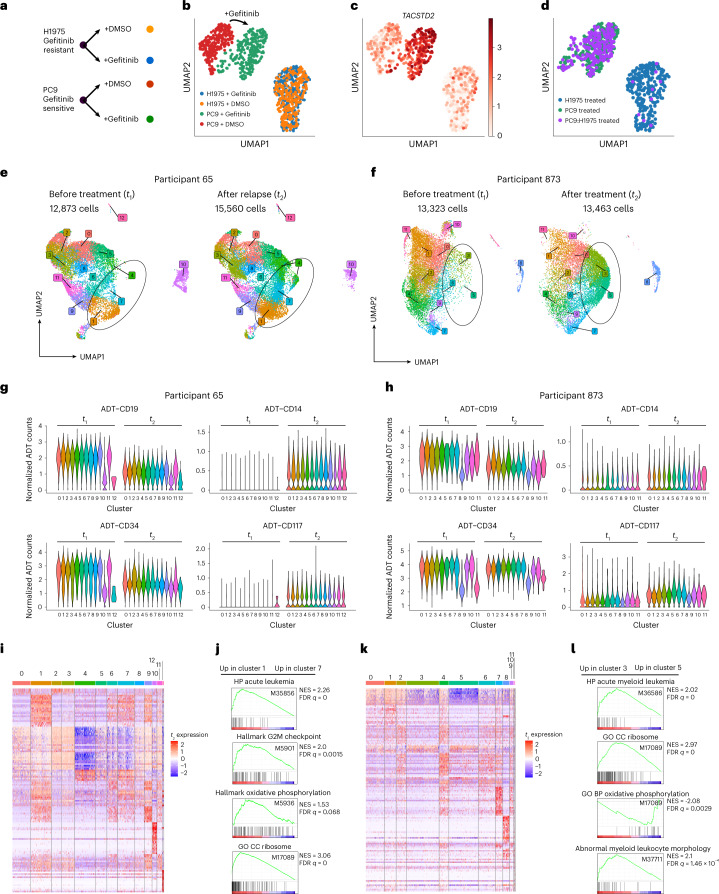
Fig. 5. Molecular signatures of drug-resistant cancer phenotypes in cell lines and human samples.
a, A two-by-two experimental study design using lung adenocarcinoma cell lines (H1975 and PC9) treated with gefitinib or DMSO. b, Clustering of scRNA-seq data after drug treatment shows transcriptional perturbations in gefitinib-sensitive PC9, but not gefitinib-resistant H1975, cells. c, Increased expression of TACSTD2 in PC9 cells challenged with gefitinib. d, Identification of drug-resistant H1975 cells spiked into drug-sensitive PC9 cells based on gefitinib-induced transcriptional perturbation. e–l, PIP-seq RNA and barcoded antibody (CITE-seq) analysis of MPAL. e, Clustering of single cells for participant 65 before (left) and after (right) chemotherapy. f, Clustering of single cells for participant 873 before (left) and after (right) chemotherapy. g,h, ADT abundance, by cluster, before (t1) and after (t2) chemotherapy. ADTs change as a function of chemotherapy but are consistent among clusters for both participant 65 (g) and participant 873 (h), with the exception of T cell subsets. i–l, Analysis of transcriptional heterogeneity in MPAL samples. i, Heat map of top differentially expressed marker genes by cluster after relapse in participant 63. j, GSEA preranked analysis comparing transcriptomic differences between clusters 1 and 7 in participant 65 using the following gene sets: Human Phenotype acute leukemia (M35856), hallmark G2M checkpoint (M5901), hallmark oxidative phosphorylation (M5936) and Gene Ontology cellular component (GO CC) ribosome (M17089). k, Heat map of top differentially expressed marker genes by cluster after relapse in participant 873. l, GSEA preranked analysis comparing transcriptomic differences between clusters 3 and 5 in participant 873 using gene sets Human Phenotype acute myeloid leukemia (M36586), Gene Ontology cellular component ribosome (M17089), Gene Ontology biological process (GO BP) oxidative phosphorylation (M17089) and abnormal myeloid leukocyte morphology (M37711).