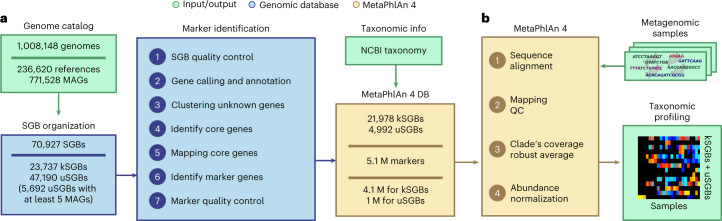
Fig. 1. MetaPhlAn 4 integrates reference sequences from isolate and metagenome-assembled genomes for metagenome taxonomic profiling.
a, From a collection of 1.01 M bacterial and archeal reference genomes and metagenomic-assembled genomes (MAGs) spanning 70,927 species-level genome bins (SGBs), our pipeline defined 5.1 M unique SGB-specific marker genes that are used by MetaPhlAn 4 (avg., 189 ± 34 per SGB). b, The expanded marker database allows MetaPhlAn 4 to detect the presence and estimate the relative abundance of 26,970 SGBs, 4,992 of which are candidate species without reference sequences (uSGBs) defined by at least five MAGs. The profiling is performed firstly by (1) aligning the reads of input metagenomes against the markers database, then (2) discarding low-quality alignments and (3) calculating the robust average coverage of the markers in each SGB that (4) are normalized across SGBs to report the SGB relative abundances (see Methods). All data are presented as mean ± s.d.