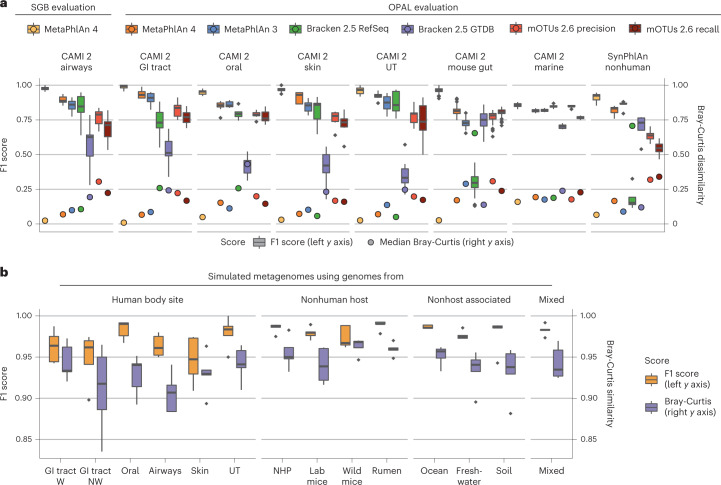
Fig. 2. MetaPhlAn 4 improves sensitivity and specificity of metagenome taxonomic profiling.
a, To evaluate its performance in taxonomic profiling, MetaPhlAn 4 was applied to synthetic metagenomes representing host-associated communities from the CAMI 2 taxonomic profiling challenge60 (n = 128 samples) and the SynPhlAn-nonhuman dataset (n = 5 samples), representing more diverse environments from previous evaluations4. Species-level evaluation using the OPAL framework61 shows that MetaPhlAn 4 is more accurate than the available alternatives in both the detection of which taxa are present (the F1 score is the harmonic mean of the precision and recall of detection) and their quantitative estimation (the BC beta-diversity is computed between the estimated profiles and the abundances in the gold standard). Additional evaluations performed using genomes within the SGB organization (labeled ‘SGB evaluation’; see Methods) show that MetaPhlAn 4 further improves accuracy at this more refined taxonomic level. See Supplementary Tables 5 and 7 for more details (GI, gastrointestinal; UT, urogenital tract). b, MetaPhlAn 4 was applied to synthetic metagenomes (n = 70 samples) modeling different host and nonhost-associated environments and containing, on average, 47 genomes from both kSGBs and uSGBs (see Methods). This evaluation directly on SGBs shows the reliability of MetaPhlAn 4 to quantify both known and unknown microbial species. Additional evaluation based on a mixture of new MAGs from samples not considered in the building of the genomic database (mixed evaluation, n = 5 samples) stresses its accuracy independently from the inclusion of the profiled data in the database. See Supplementary Tables 9 and 10 for more details (NHP = nonhuman primates, W = westernized, NW = nonwesternized). Box plots in a and b show the median (center), 25th/75th percentile (lower/upper hinges), 1.5× interquartile range (whiskers) and outliers (points).