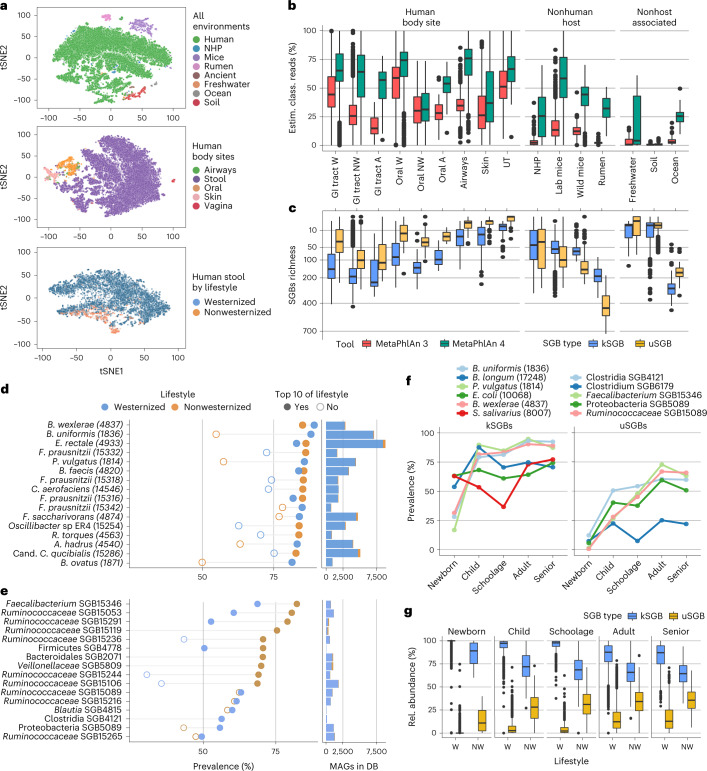
Fig. 3. MetaPhlAn 4 expands observable microbial diversity, primarily by quantifying yet-to-be-characterized species (uSGBs).
a, We applied MetaPhlAn 4 profiling to a total of 24.5 k metagenomic samples from diverse environments, highlighting its ability to detect microbiome compositions and clear differences between them, even when considering distinct human body sites and variable host lifestyles (Supplementary Fig. 5b and Supplementary Table 11). b, The expanded genomic database of MetaPhlAn 4 substantially increases the estimated fraction of classified reads in comparison with the previous MetaPhlAn version across habitat types (n = 24,515 samples). c, MetaPhlAn 4 detects on average 48 unknown bacterial species (uSGBs) per human gut microbiome, and reaches up to more than 700 in other nonhuman environments (n = 24,515 samples). d, The most prevalent microbial species in the gastrointestinal tract of westernized populations are known species (kSGBs). The ten most prevalent kSGBs in westernized and nonwesternized lifestyles are shown ordered by their highest prevalence and reported together with the number of MAGs assembled from human gut metagenomes in the MetaPhlAn genome catalog. Species names are shown together with their SGB ID between brackets. e, The most prevalent SGBs in nonwesternized populations belong to yet-to-be-cultivated and named species. The ten most prevalent uSGBs of each lifestyle are shown ordered by their highest prevalence. f, In westernized populations, the most prevalent kSGBs and uSGBs vary across age categories. The two most prevalent SGBs for each age category are shown. g, The fraction of uSGBs relative to kSGB increases after infancy (n = 19,468). Box plots in b, c and g show the median (center), 25th/75th percentile (lower/upper hinges), 1.5× interquartile range (whiskers) and outliers (points). NHP, nonhuman primates; W, westernized; NW, nonwesternized; A, ancient.