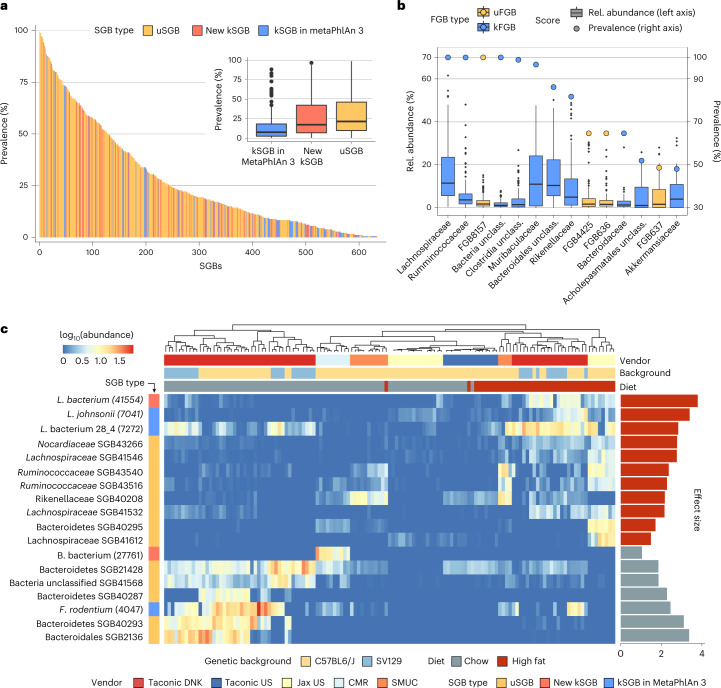
Fig. 4. MetaPhlAn 4 enables accurate metagenomic profiling of mouse microbiomes containing few cultured isolate taxa.
a, MetaPhlAn 4 taxonomic profiling of a cohort of mouse gut microbiome samples (n = 181 samples), spanning eight genetic backgrounds and six different vendors65 revealed that the majority of detected microbial taxa are uncharacterized SGBs (uSGBs) that do not contain a sequenced isolate representative. b, Some of the most prevalent families in the mouse gut microbiome (n = 181 samples) are still unclassified at the family level (uFGBs). FGBs detected in at least 20% of the samples (circles and right-side y axis) and with a median relative abundance above 1% (box plots and left-side y axis) are shown. c, Random effects models applied to the MetaPhlAn 4 profiles revealed that most of the high- and low-fat diet microbial biomarkers are uncharacterized species (FDR < 0.2). log10-transformed relative abundances of the microbial biomarkers are represented in the heatmap and their effect size (linear model beta coefficient) in the bar plots. For kSGBs, species names are shown together with their SGB ID between brackets. SGB41568 is reported in NCBI as assigned to an unclassified phylum, and we thus report only the kingdom label. SMUC = Southern Medical University in China, CMR = Craniofacial Mutant Resource at the Jackson Laboratory (Jax). Box plots in a and b show the median (center), 25th/75th percentile (lower/upper hinges), 1.5× interquartile range (whiskers) and outliers (points).