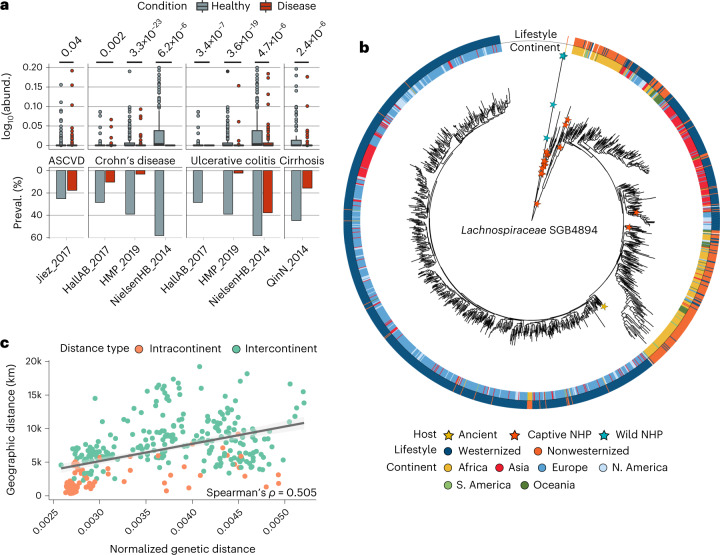
Fig. 6. StrainPhlAn 4 accurately reconstructs large-scale strain-level phylogenies of uncharacterized microbial species.
a, Relative abundances (box plots and top-part y axis) and prevalences (bar plots and bottom-part y axis) of the uncharacterized species (uSGB) Lachnospiraceae SGB4894 are substantially higher in healthy individuals (n = 738 samples) in comparison with patients suffering from several gastrointestinal related diseases (n = 1,183 samples), and this difference is reproducible across populations (one-sided Mann–Whitney U test). Box plots show the median (center), 25th/75th percentile (lower/upper hinges), 1.5× interquartile range (whiskers) and outliers (points). b, Lachnospiraceae SGB4894 shows within-species genetic diversity strongly linked to geographic origin and lifestyle. c, Pairwise geographic distances between strains of different countries correlate with their median genetic distances (Spearman’s ρ = 0.505; see Methods), suggesting that human Lachnospiraceae SGB4894 strains could have followed an isolation-by-distance pattern.