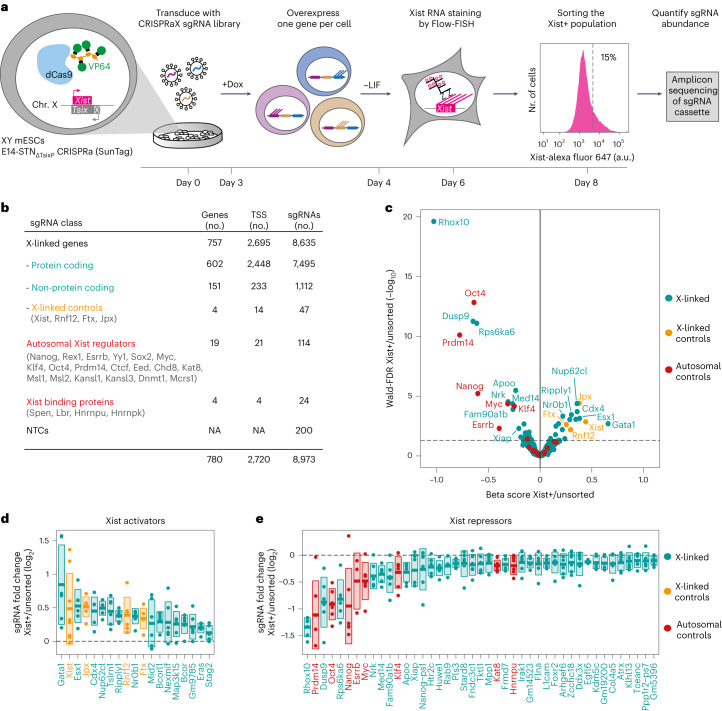
Fig. 1. Pooled CRISPR activation screen identifies unknown Xist regulators.
a, Schematic depiction of the CRISPRa screen workflow. A male ESC line with a deletion of the major Tsix promoter and a stably integrated doxycycline-inducible CRISPRa SunTag system (E14-STNΔTsix) was transduced with a custom sgRNA library targeting X-chromosomal genes (CRISPRaX). Following puromycin selection, the cells were treated with doxycycline (Dox) to overexpress one gene per cell, and differentiated by LIF withdrawal (–LIF) to induce Xist upregulation. Cells were stained with Xist-specific probes by Flow-FISH and the top 15% Xist+ cells were sorted by flow cytometry. The sgRNA cassette was amplified from genomic DNA and sgRNA abundance in the unsorted and sorted populations was determined by deep sequencing. The screen was performed in three independent replicates. b, Composition of the CRISPRaX sgRNA library, targeting each TSS with six sgRNAs per gene. Because a subset of guides target multiple coding and non-coding transcripts, the total number of sgRNAs is smaller than the sum of sgRNAs across categories. c, Volcano plot of the screen results, showing the beta-score as a measure of effect size versus Wald-FDR (MAGeCK-MLE), coloured according to gene class. The dotted line denotes Wald-FDR < 0.05. d,e, Comparison of individual sgRNA abundance (dots) in the sorted fraction compared with the unsorted population for all significantly enriched (d) or depleted (e) genes in the screen (Wald-FDR < 0.05, MAGeCK-MLE). The mean of three independent replicates is shown. Genes are ordered by their beta-score, a measure for effect size (MAGeCK-MLE). The central line depicts the mean, boxes depict the standard deviation across all sgRNAs targeting the respective gene. Only the highest scoring TSS per gene is depicted. Source numerical data are available as source data.