Figure 3.
LEEs are highly prevalent and diversifies with the main chromosome
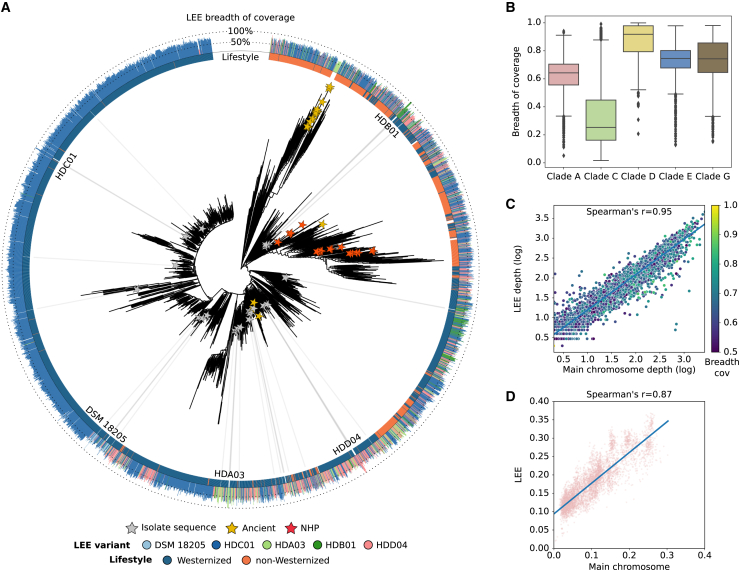
(A) Phylogenetic reconstruction of S. copri (clade A). The length of the external bar plots represents the breadth of coverage of LEE in the sample. NHP, non-human primate.
(B) Breadth of coverage of LEE when the depth of coverage of the main chromosome is above 1×.
(C) Spearman correlation between the depth of coverage of the main chromosome (x axis) and LEE (y axis) of S. copri (clade A), when the depth of coverage of the main chromosome is above 2× and the breadth of coverage of LEE is above 50% (Spearman’s r = 0.95). The color gradient represents the breadth of coverage of LEE.
(D) Spearman correlation of the single-nucleotide polymorphisms (SNPs) rates between the main chromosome (x axis) and LEE (y axis) of S. copri (clade A), when the breadth of coverage of LEE is above 80% (Spearman’s r = 0.87). SNP rates of the main chromosome were calculated using the multiple-sequence alignment (MSA) of the StrainPhlAn marker genes and those from LEE using the MSA of the full LEE alignment. Boxplots in (B) show the median (center), 25th/75th percentile (lower/upper hinges), 1.5× interquartile range (whiskers), and outliers (points). See also Figures S2–S5.