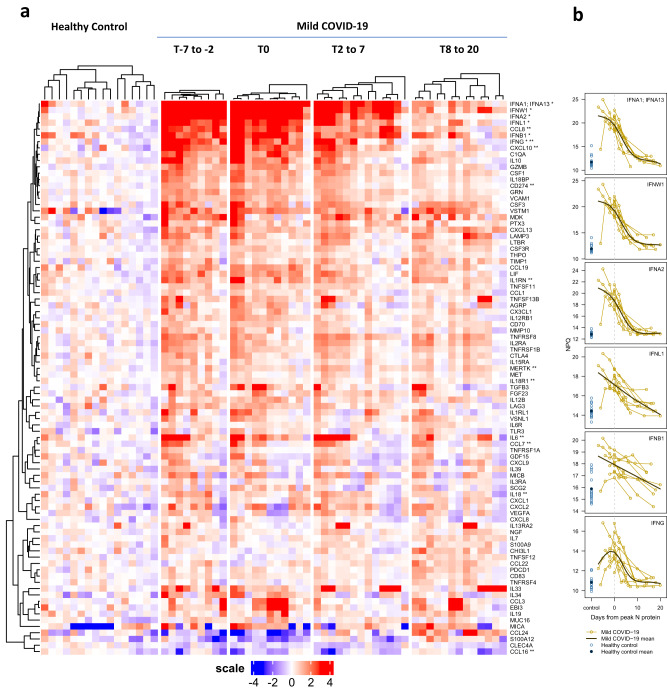
Fig. 5. Heatmap of differential protein abundance between SARS-CoV-2-infected patients and healthy controls at different time points.
T0 (n = 9) represents time of peak expression of SARS-CoV-2 nucleocapsid protein (N-protein); T-7 to -2 (n = 11) represents 2–7 days before T0, T2 to 7 represents 2–7 days after T0 (n = 13), and T8 to 20 represents 8–20 days after T0 (n = 13). Mixed effect linear model analysis was performed for each time point comparing COVID-19 samples to healthy controls (n = 16). a A total of 88 differential abundance proteins were identified and visualized in a clustered heatmap. The heatmap displays the log2-normalized read counts (NULISA Protein Quantification, NPQ) centered relative to the mean in the healthy control samples; values above the mean are shown in red and values below the mean are shown in blue. Target names provided in the text are marked with asterisks: * indicates interferons, and ** indicates targets previously reported to be associated with COVID-19. b Individual trajectories of interferon abundance (NPQ) relative to days from peak N-protein are shown for the mild COVID group (gold lines). Trajectories are aligned based on the day of peak N-protein (dashed gray vertical line). Generalized additive models with a cubic regression spline basis were used to estimate the mean mild COVID-19 trajectory (dark brown line). Control samples (lighter blue open circles) and control group mean (dark blue solid circle) are shown to the left. Source data are provided in the Alamar_NULISAseq_COVID_NPQ.csv file available under accession GSM7734324.