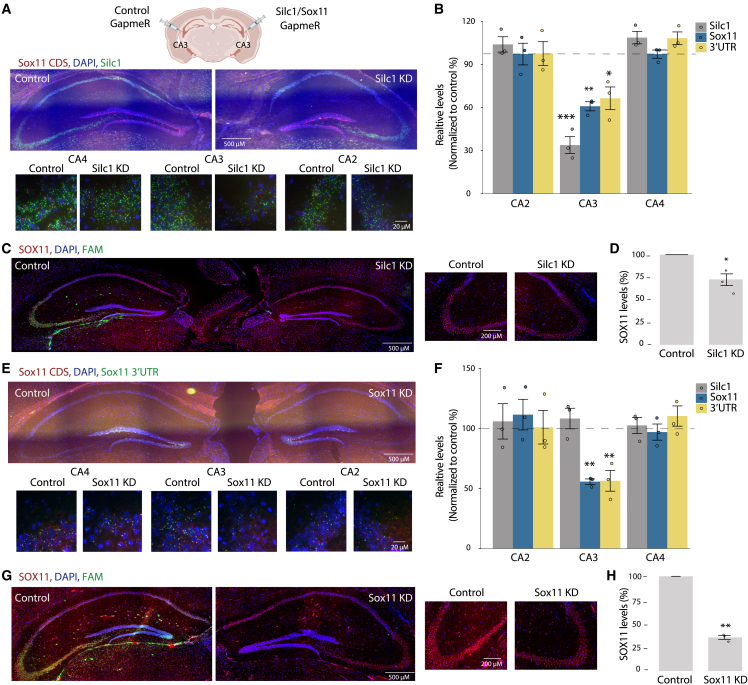
Figure 3.
Silc1 and Sox11 KD by injections of GapmeRs to the CA3 region
(A) Control Fluorescein amidites (FAM)-labeled (left) or Silc1-targeting (right) GapmeRs were injected into the CA3 region. 5 days later, the hippocampus was extracted for coronal sections. RNAscope analysis of Silc1 and Sox11 expression was performed using Silc1 (green) and Sox11 CDS (red) probes and DAPI. Imaging was done using 20× (scale bar, 200 μm) and 100× oil immersion objectives (scale bar, 20 μm).
(B) RNAscope quantification of the number of green and red dots, normalized to control GapmeR, performed using IMARIS software. 12 images of non-overlapping fields were quantified per biological repeat; 3 biological repeats. Mean ± SEM is shown. The p value was calculated using an unpaired two-sample t test; ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.001.
(C) Immunostaining with anti-SOX11 (red) and DAPI (blue) in hippocampi of Silc1 KD mice. The FAM signal marks the injection site of the control GapmeR. Imaging was performed using a 20× objective (scale bar, 200 μm).
(D) Quantification of 3 biological repeats of hippocampus staining, normalized to injection of control GapmeR. Mean ± SEM; ∗p < 0.05, unpaired two-sample t test.
(E) As in (A) using control (left) or Sox11 CDS (right) GapmeRs.
(F) As in (B) using control or Sox11 CDS GapmeRs.
(G) As in (C) for Sox11 KD mice.
(H) As in (D) for Sox11 KD mice.
See also Figure S3.