Figure 1.
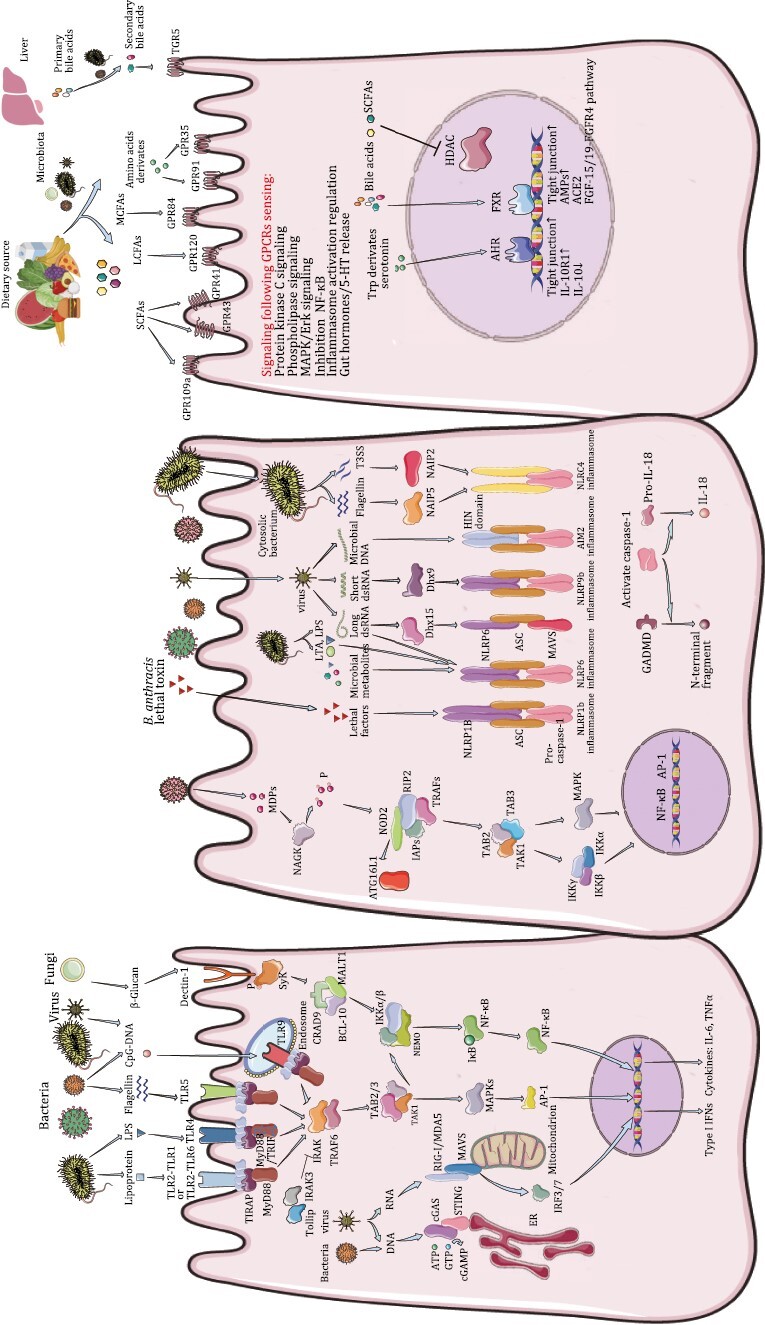
Microbial signal–host sensor interactions in IECs and associated signaling events. (Left) Microorganism-associated molecular patterns (MAMPs) from the bacteria, fungi and viruses are sensed by extensive types of pattern recognition receptors (PRRs), which are broadly expressed in/on intestinal epithelial cells (IECs). For toll-like receptors (TLRs), TLR4 senses bacterial lipopolysaccharide (LPS), TLR2-TLR1 or TLR2-TLR6 dimers sense bacterial lipoprotein, TLR5 senses bacterial flagellin and TLR9, which mainly localized on endosome membrane, is triggered by CpG DNA from both bacteria and viruses. Once binding ligands, TLR homologous or heterodimers are formed and MyD88-dependent and/or MyD88-independent signaling pathways are activated by different adaptor proteins (AP) such as MyD88, Toll/IL-1 receptor (TIR) domain-containing adaptor protein (TIRAP), TIR domain-containing adaptor molecule (TRAM) and/or TIR domain-containing adaptor inducing IFNβ (TRIF). However, TLR2 and TLR4 need TIRAP to recruit MyD88. TLR4 can initiate downstream signals using both MyD88 and TRIF as signaling adaptors, but it also needs TRAM to recruit TRIF and TIRAP to recruit MyD88. MyD88 attracts IL-1 receptor-associated kinases (IRAKs) to TLRs through the interaction of two molecular death domains and IRAK-1 is activated by phosphorylation, which then interacts with TNF-receptor-associated factor (TRAF), a ubiquitin ligase. Typically, TRAF6 forms complexes with TGF-beta activated kinase (TAK) and TAK1-associated binding protein 2 (TAB), and TAK activates downstream mitogen activated protein (MAP) kinase [Jun N-terminal kinases (JNK), p38 MAPK] and inhibitor of nuclear factor kappa-B kinase (IKK), leading to activation of nuclear factor kappa-B (NF-κB) and activator protein 1 (AP-1), and therefore regulating the expression of pro-inflammatory cytokines and other immune-related genes. TLR4 and TLR3 could induce TRIF-dependent IFN-I pathway, and activate similar downstream signals through TRAF3 or receptor-interacting serine/threonine-protein kinase 1 (RIPK1)/TRAF6. Both Tollip and IRAK-3 interact with IRAK-1 and negatively regulate TLR-mediated signal transduction pathways. Viruses are sensed by retinoic acid-inducible gene I (RIG-I), which detect 5ʹ-ppp RNA and short-chain dsRNA, and melanoma differentiation-associated gene 5 (MDA5), which detect long-chain dsRNA respectively, and lead to type I IFN production in a mitochondrial antiviral signaling protein (MAVS)- and stimulator of interferon response cGAMP interactor (STING)-dependent manner. DNA viruses activate the cyclic GMP-AMP synthase (cGAS)-STING pathway. In the presence of DNA, cGAS enzymatically converts ATP and GTP into the cytosolic dinucleotide, cGAMP, which then binds to STING and activates IFN signaling. Dectin-1, officially named as C-type lectin domain-containing 7A (CLEC7A), a member of the C-type lectin family, senses β-glucans from fungi and promotes spleen tyrosine kinase (SyK) phosphorylation and subsequent signaling activation. (Mid) NLRs are a subset of cytoplasmic PRRs. On sensing their respective peptidoglycan ligands [d-glutamyl-meso-diaminopimelic acid (DAP) or phosphorylated muramyl dipeptide (MDP)], nucleotide-binding oligomerization domain-containing 1 (NOD1) and NOD2 undergo auto-oligomerization and induce NF-κB signaling by activating receptor-interacting serine/threonine kinase 2 (RIP2). Or NODs can interact with autophagy related 16 like 1 (ATG16L1) to induce autophagy. ATG16L1 is also a negative regulator of NOD signaling. NOD-like receptors (NLRs) such as NLRP1b, NLRP6 and NLRP9b oligomerize upon activation to form inflammasome complexes, which serve as platforms for the recruitment, cleavage, and activation of the inflammatory Caspase-1 (CASP-1). Subsequently, activated CASP-1 can process pro-IL-1β, pro-IL-18 or gasdermin D (GSDMD) into active forms, which in turn stimulate cytokine secretion or pyroptosis. Furthermore, NLRP6 can sense and bind the long viral dsRNA directly or through the RNA helicase DEAH-box helicase 15 (DHX15), which forms liquid–liquid phase separation (LLPS) to integrate inflammasome and and ISG. Microbial metabolites such as SCFAs could regulate NLRP6 inflammasome function. NLRP9b senses and binds the short viral dsRNA through DHX9 and activates the inflammasome. Flagellin and PrgJ, a conserved type III secretion system (TTSS) rod component from bacteria, bind to their corresponding adaptors, NAIP5 and NAIP2, respectively. Subsequently, they interact with NLRC4 to initiate formation of the NLRC4 inflammasome. (Right) Microbiota-processed metabolites could be sensed mainly by G Protein-Coupled Receptors (GPCRs) on cell membrane or by aryl hydrocarbon receptor (AHR) and nuclear receptors in cell nucleus and induce downstream signaling pathways. GPR41/43/109a are part of typical receptors for short-chain fatty acids (SCFAs, <6C), GPR120 could sense long -chain fatty acids (LCFAs, >12C) and GPR84 is stimulated by medium-chain fatty acids (MCFAs, 6C~12C). SCFAs could directly inhibit the activity of Histone deacetylase (HDAC). Amino acids derivates such as tryptophan and tyrosine catabolism products could activate GPR35, and succinate binds GPR91. Tryptophan derivatives, for example, indole-3-aldehyde could also stimulate nuclear AHR. Primary bile acids are produced in liver and some of them would be metabolized into secondary bile acids by microbiota when come to distal intestine. Nuclear receptor farnesoid X-activated receptor (FXR) can bind bile acids and regulate bile acid metabolism. G protein-coupled bile acid receptor 1 (GP-BAR1, also named TGR5) is a membrane GPCR and mainly binds to secondary bile acids.
