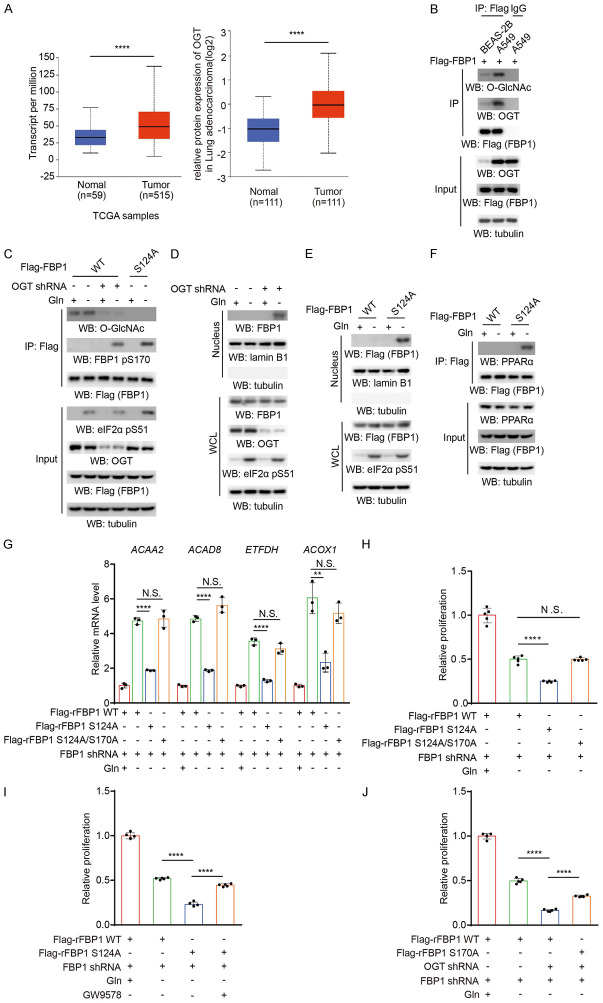
Figure 3.
O-GlcNAcylation inhibits the nuclear translocation of FBP1 and promotes β-oxidation gene expression in NSCLC cells. A. Left: A TCGA dataset was used to analyze OGT expression levels in normal lung tissues and LUAD tissues. Normal samples: minimum, 9.876; maximum, 76.777; and median, 33.21; LUAD samples: minimum, 5.37; maximum, 138.023; and median, 48.894. The horizontal lines mark the median. ****P < 0.0001. Right: The CPTAC dataset was used to analyze OGT protein levels in normal lung tissues and LUAD tissues (https://ualcan.path.uab.edu/analysis-prot.html). Normal samples: minimum, -2.722; maximum, 0.317; and median, -1.026; LUAD samples: minimum, -2.014; maximum, 2.103; and median, -0.032. The horizontal lines mark the median. ****P < 0.0001. B. BEAS-2B and A549 cells expressing Flag-FBP1 were harvested for immunoprecipitation analysis with an anti-Flag antibody. Immunoblotting analyses were performed with the indicated antibodies. C. The indicated Flag-FBP1 proteins were expressed in A549 cells with or without OGT shRNA expression, which were treated with or without glutamine deprivation for 6 h. Immunoprecipitation analyses were performed with an anti-Flag antibody. Immunoblotting analyses were performed with the indicated antibodies. D. A549 cells with or without OGT shRNA expression were treated with or without glutamine deprivation for 6 h. Nuclear fractions were prepared. Immunoblotting analyses were performed with the indicated antibodies. E. The indicated Flag-FBP1 proteins were expressed in A549 cells, which were treated with or without glutamine deprivation for 6 h. Nuclear fractions were prepared. Immunoblotting analyses were performed with the indicated antibodies. F. The indicated Flag-FBP1 proteins were expressed in A549 cells treated with or without glutamine deprivation for 6 h. Immunoprecipitation analyses were performed with an anti-Flag antibody. Immunoblotting analyses were performed with the indicated antibodies. G. A549 cells with depleted endogenous FBP1 and reconstituted expression of the indicated Flag-rFBP1 proteins were treated with or without glutamine deprivation for 12 h. The relative mRNA levels of the indicated genes were determined. The data represent the mean ± SD of 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by two-tailed Student’s t test; N.S.: not significant by two-tailed Student’s t test. H. A549 cells with depleted endogenous FBP1 and reconstituted expression of the indicated Flag-rFBP1 proteins were treated with or without glutamine deprivation for 48 h. Cell viability was measured by CCK-8 assays. The data represent the mean ± SD of 5 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by two-tailed Student’s t test; N.S.: not significant by two-tailed Student’s t test. I. A549 cells with depleted endogenous FBP1 and reconstituted expression of the indicated Flag-rFBP1 proteins were treated with or without glutamine deprivation and GW9578 (500 nM) for 48 h. Cell viability was measured by CCK-8 assays. The data represent the mean ± SD of 5 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by two-tailed Student’s t test; N.S.: not significant by two-tailed Student’s t test. J. A549 cells with depleted endogenous FBP1 and reconstituted expression of the indicated Flag-rFBP1 proteins with or without OGT shRNA expression were treated with or without glutamine deprivation for 48 h. Cell viability was measured by CCK-8 assays. The data represent the mean ± SD of 5 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by two-tailed Student’s t test; N.S.: not significant by two-tailed Student’s t test.