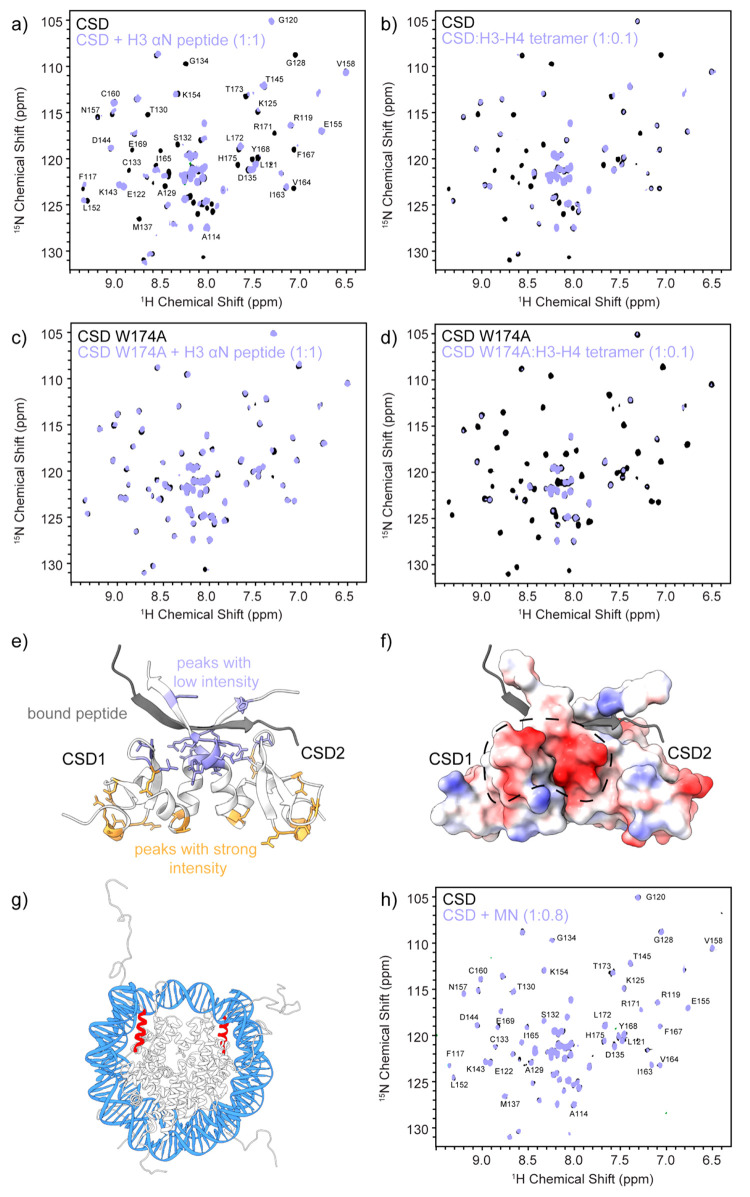
Figure 2.
Interactions of the CSD dimer with H3 and the nucleosome. 2D HSQC experiments of a sample prepared with (a) 15N-labeled CSD dimer and natural abundance H3(37–59) peptide in a 1:1 ratio, (b) 15N-labeled CSD dimer and natural abundance H3–H4 tetramer in a 1:0.1 ratio, (c) 15N-labeled CSD W174A dimer and natural abundance H3(37–59) peptide in a 1:1 ratio, and (d) 15N-labeled CSD W174A dimer and natural abundance H3–H4 tetramer in a 1:0.1 ratio. (e) Structure of the CSD dimer with bound peptide (gray). Residues corresponding to peaks that lose intensity upon addition of the H3(37–59) peptide as determined in part a are shown in purple, while residues that retain their intensity are depicted in orange. (f) Electrostatic map of the CSD dimer illustrating a negatively charged patch that can interact with histone proteins through nonspecific interactions. (g) Structure of the nucleosome depicting the position of H3(37–59) shown in red. (h) 2D HSQC experiment of a sample containing 15N-labeled CSD dimer and natural abundance mononucleosomes in a 1:0.8 ratio. See Figure S7 for intensity ratio analysis.