Fig. 5.
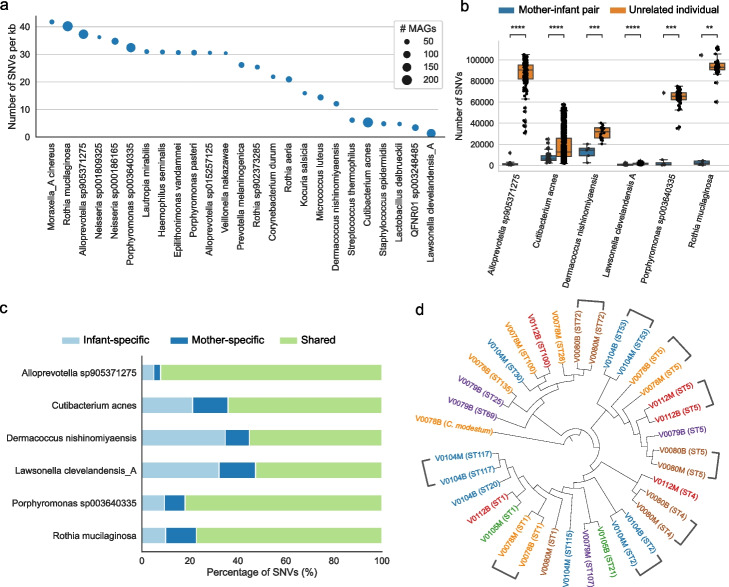
Single-nucleotide variation indicates mother–infant microbial sharing. a Top species with the largest intraspecies SNV density. The size of dots indicates the number of MAGs corresponding to each species. b Number of SNVs in pairwise comparisons between mother-infant pairs and between infants and unrelated mothers. Only species with genomes from at least four mother–infant pairs were considered for analysis. Statistical significance was tested by two-tailed Wilcoxon rank sum test. **P < 0.01, ***P < 0.001, ****P < 0.0001. c Proportion of SNVs that were found in genomes from infants only or mothers only or both. SNVs were called based on the species-level representative MAG as the reference genome. d Phylogenetic tree of representative C. acnes cultured isolates with C. modestum as the outgroup. Source of individual is indicated in the label name and label color. Sequence type is displayed in parentheses