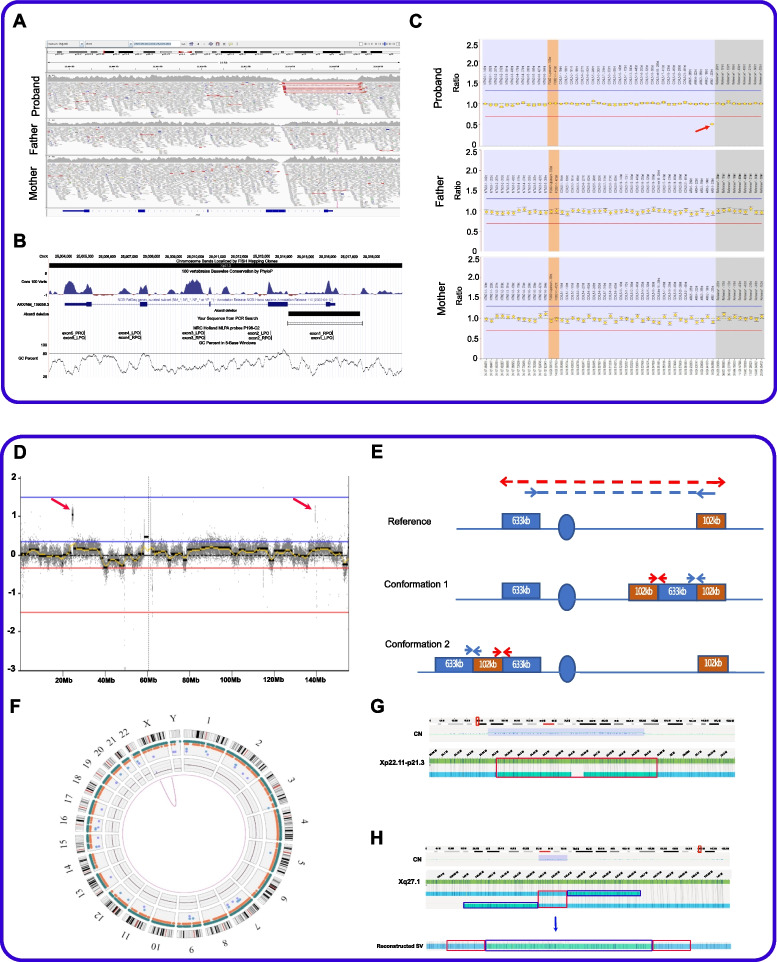
Fig. 3.
Validation data for two patients with X-linked structural variants in OxClinWGS study. A–C Patient with ARX deletion: A screenshot showing 125 bp read alignments supporting a de novo deletion of ARX exon 1. Region shown is chrX:25,003,000–25,019,000 (GRCh38). Visualisation is using IGV v2.11.2, with squished and “view as pairs” options. Alignments are coloured by insert size and transcript shown is NM_139058.3. B UCSC genome browser session showing the position of the deletion in relation to the PCR primers and MLPA probes that were used for validation. Also shown is the GC content which rises to > 80% near the distal breakpoint where the coverage drops also in parental genomes. An interactive version is available at https://genome.ucsc.edu/s/AlistairP/ARX_deletion_v6. C The 3 kb deletion was confirmed by the MLPA validation data visualised using coffalyser and shows a drop in signal only for the proband for the exon 1 probe (red arrow). The grey boxes are reference probes and the orange boxes highlight the 95% confidence range of the reference samples used. D–H Complex rearrangement in patient with X-linked neurodevelopmental disorder. D Read count information from short-read sequencing normalised by ngCGH software (https://github.com/seandavi/ngCGH) showing two X chromosome duplications (red arrows). E Split Illumina read-pairs suggest the two duplications are inter-linked. However, two possible configurations can explain the split read pattern. F Circos plot highlights the only SV identified by the Bionano pipeline above the threshold for SV detection. G Genome browser view of the optical maps robustly detects the ~ 600 kb duplication of Xp22.11p21.3 being inserted into Xq27.1, which is present in the carrier mother and both affected male siblings using the Bionano pipeline excluding Complex Multi-Path Regions (CMPR). The red box highlights the duplication inserted into Xq27.1. H The Bionano pipeline without masking CMPR detects ~ 102 kb tandem duplication (red boxes) flanking either side of the 600 kb insertion from Xp22.11-Xp21.3 (blue box), therefore, supporting conformation 1 as suggested in E