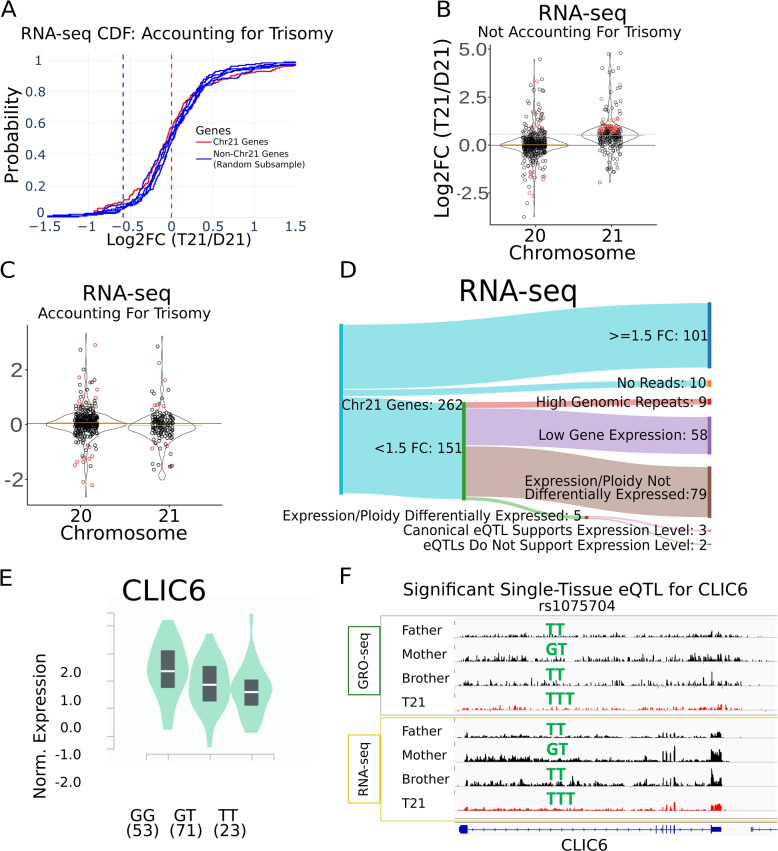
Fig. 3.
Alternative explanations to disparate fold change estimates. A Cumulative distribution plot of fold changes found in real RNA-seq data, after accounting for trisomy. The solid red line indicates all chromosome 21 genes. Each solid blue line is a randomly selected set of genes from all other chromosomes. B Violin plots indicating Log2 fold change between T21 and D21 samples. Significant gene calls are colored red (padj ). C Same as B, but using a trisomy-aware pipeline for analysis. D Sankey diagram depicted the filtering process of our RNA-seq analysis. The initial 151/262 genes identified as potentially dosage compensated can alternatively be explained by genomic repeats, high variance from low expression genes, or technical artifacts related to failing to normalize the data to the ploidy number. The remaining genes can be explained by the presence of eQTLs (see also Additional File 1: Fig. S22)). E Example boxplot indicating relative expression of the gene CLIC6 with one eQTL. F Genome viewer tracks for the gene CLIC6 for all four family members, in GRO-seq (top) and RNA-seq (bottom). The T21 track is indicated in red. The allelic makeup of the eQTL in E is indicated by the green text above each track