Abstract
Background
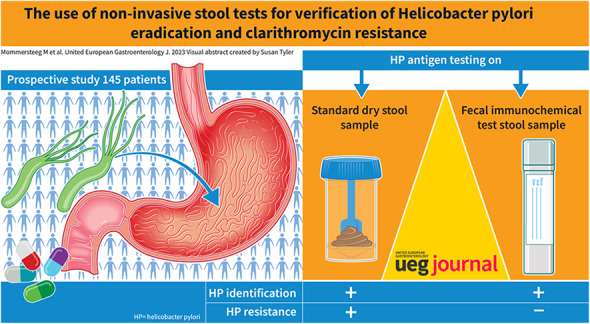
Clarithromycin resistance of Helicobacter pylori (H. pylori) represents a major challenge in eradication therapy. In this study, we assessed if non‐invasive stool tests can be used to verify successful H. pylori eradication and determine clarithromycin resistance.
Materials and methods
In this prospective study, patients undergoing urea breath testing (UBT) for confirmation of H. pylori eradication were asked to collect the stool as both a dry fecal sample and fecal immunochemical test (FIT). Stool H. pylori antigen testing (SAT) was performed on these samples and assessed for its accuracy in eradication verification. Type and duration of antibiotic treatment were retrospectively collected from patient records and compared with clarithromycin resistance determined by PCR of stool samples.
Results
H. pylori eradication information was available for a total of 145 patients (42.7% male, median age: 51.2). Successful eradication was achieved in 68.1% of patients. SAT on FIT samples had similar accuracy for eradication assessment compared to dry fecal samples, 72.1% [95% CI 61.4–81.2] versus 72.2% [95% CI 60.9–81.7]. Clarithromycin resistance rate was 13.4%.
Conclusion
H. pylori antigen testing on FIT stool samples to verify H. pylori eradication is feasible and has similar accuracy as H. pylori antigen testing on dry stool samples. Dry stool, but not FIT, was suitable for non‐invasive identification of H. pylori clarithromycin resistance by rt‐PCR personalizing antibiotic treatment strategies without the need for invasive diagnostics is desirable, as the cure rate of first‐line empirical H. pylori treatment remains low.
Keywords: antibiotic resistance, clarithromycin, Faecal Immunochemical Test (FIT), gastric cancer, H. pylori eradication therapy, Helicobacter pylori, stool antigen test, urea breath test
Key summary.
Summarize the established knowledge on this subject
H. pylori fecal antigen testing can be performed on stool collected by FIT.
H. pylori eradication remains challenging.
Clarithromycin resistance of H. pylori represents the main challenge in H. pylori eradication therapy.
What are the significant and/or new findings of this study?
Empirical eradication treatments have limited success rates, with an eradication rate of 68%.
We showed that H. pylori antigen testing on dry fecal stool and on stool collected by FIT is comparable to assess H. pylori eradication, with an accuracy of 70%.
H. pylori resistance patterns can be accurately determined on solid stool, but not on FIT by means of rt‐PCR.
INTRODUCTION
Infection with the rod‐shaped gram‐negative bacterium Helicobacter pylori (H. pylori) poses the main risk factor for gastric adenocarcinoma (gastric cancer). 1 Treatment of H. pylori significantly reduces the risk of gastric cancer. Therefore, H. pylori eradication is widely recommended, with some high‐risk areas even considering a search and treat strategy. 2 The European Union has recently recommended intensifying population screening for cancer and to include gastric cancer screening under certain circumstances. 3 An emerging challenge in the treatment of H. pylori is the increasing incidence of antibiotic resistance. Evidence suggests that within Europe, resistance rates are rising and may reach up to 30%. 4 The Maastricht/Florence guideline on the management of H. pylori advises susceptibility testing before starting antibiotic treatment if the regional clarithromycin resistance is above 15%. This threshold was reached in 13 out of 18 investigated European countries in 2018, including the Netherlands. 5 , 6 , 7 , 8 Susceptibility testing is traditionally performed by bacterial culture and development of an antibiogram. An obvious disadvantage to this diagnostic approach is the requirement of fresh biopsies. 9 , 10 Another challenge is the culture of H. pylori itself, which even in experienced hands can be demanding and time‐consuming. Newer molecular methods have been developed to investigate H. pylori resistance, most comprising a real‐time polymerase chain reaction (rt‐PCR) detecting mutations in the 23S ribosomal rRNA gene of the bacteria—mutations which prevent the binding of macrolide‐type antibiotics, including clarithromycin, rendering these bacteria resistant to such treatments. 11 , 12 This mechanism is relevant since clarithromycin resistance of H. pylori appears to represent the main challenge in H. pylori eradication in Europe, where most first‐line empiric therapies include this antibiotic. 7 While PCR‐based techniques may eliminate the need for microbial culture and thereby assess antibiotic resistance more robustly, a gastric biopsy is still needed to perform these procedures. As gastroduodenoscopy is an invasive test associated with considerable burden, risks and healthcare costs, the need for patients to undergo an endoscopy for diagnosis and treatment currently presents a significant drawback for both molecular and culture‐based methods. 10
To implement H. pylori diagnosis and antibiotic resistance testing at higher volumes, non‐invasive tests are highly preferable. In recent years, PCR‐based approaches have been evaluated on patient fecal material with promising results; however, this method has not yet reached the main clinical practice. 13 , 14 , 15 , 16 , 17 We have previously shown that the non‐invasive stool antigen test (SAT), which detects H. pylori antigen in fecal matter, is as efficient for the diagnosis of H. pylori infection as the urea breath test (UBT). This study also showed that patients are more comfortable collecting fecal immunohistochemical test (FIT) samples than solid fecal material 18 and that fluid collected from FIT tests was equally effective for H. pylori stool antigen detection as solid stool samples.
Thus, here we investigated to what extent H. pylori antigen tests in FIT fluid and solid feces may be used to verify the efficacy of eradication treatment. As a secondary aim, we attempted to assess antibiotic resistance in collected stool samples.
MATERIALS AND METHODS
Study design
The design of this study has been published previously. 18 In short, this prospective study was performed in two hospitals (one academic and one regional) in the Netherlands between February 2018 and December 2020. Patients were eligible for inclusion if they were over 18 years of age and were referred for C13 urea breath testing (UBT) for either primary diagnosis of H. pylori infection or verification of eradication. Patients were excluded if they had used antibiotics/bismuth in the past 4 weeks, or a proton pomp inhibitor (PPI) in the past 2 weeks. All participants underwent UBT, and were instructed to collect both a dry stool and a FIT sample before the UBT took place (see details below). Results of the UBT were considered as reference. For the current study, the history of H. pylori treatment and outcome thereof were collected from electronic patient files. For the PCR experiments a sub‐selection of patients was analyzed, consisting of all patients testing positive for UBT for whom fecal DNA was available, and an equal number of (randomly selected) UBT‐negative patients. See Figure 1 for a flow chart of inclusion to the current study. The institutional review boards of both participating hospitals approved the study (MEC‐2017‐528). This trial was registered in the Dutch trial register (NTR7052) and was supported by the Dutch Digestive Foundation (D18‐02).
FIGURE 1.

Flowchart of patient inclusion in this study. The HELI cohort comprises 182 individuals invited for UBT for either a primary diagnosis of Helicobacter pylori (H. pylori) or eradication thereof. We retrieved electronic patient records on H. pylori eradication for 145 of these patients, 88 of whom underwent UBT for verification of treatment efficacy and had stool and/or FIT samples available for stool antigen measurements of these patients. For the detection of antibiotic resistance using rt‐PCR, stool samples were obtained from 97 patients with and without H. pylori infection from the original HELI cohort (including those with primary infections and those tested for eradication of H. pylori infection). UBT, urease breath test.
Fecal sampling
Feces sampling for both dry stool and FIT samples was performed by participants at home. Samples were collected within 24 h before the scheduled UBT, at least 2 weeks after the completion of their H. pylori eradication treatment. The dry stool was collected in a standard feces container (DeklaPack Europe) without any medium inside. Using a small shovel attached to the lid of the collection tube, patients deposit a small clump of feces into the container. The FIT collection tube that was used was the FOB Gold (Sysmex). The lids of these tubes, in which a sample buffer is present, are provided with a sampling stick with notches. Dipping the stick in the feces collects a (small) standardized amount of fecal material, which is subsequently deposited in the buffer inside the FIT collection tube.
Fecal antigen ELISA
For the fecal antigen ELISA, a commercial kit was used (Fecal Helicobacter pylori Antigen, ref KT 826, Epitope Diagnostics Inc.). All tests were performed according to the manufacturer's guidelines. In short, 40 mg of fecal material was suspended in 1 mL of assay buffer and mixed. Of this sample, 100 µL was added to monoclonal antibody‐coated microwell plates, which were incubated for 60 min. The wells were washed and the tracer antibody was added and incubated for 30 min. Then, the wells were again washed and the HRP substrate was added. Quantification was performed using an Infinite M Nanoplatereader (Tecan Group Ldt.) at a wavelength of 450 nm.
To measure fecal H. pylori antigens in FIT, the same protocol was used, with minor modifications. Instead of feces, 1 mL of FIT liquid was centrifuged for 1 min at 14,000 g, and 100 µL of the supernatant was used for the assay without the addition of assay buffer. As an extra negative control, unused FIT fluid was measured, which gave similar readings to assay buffer without fecal material.
Real time PCR for clarithromycin resistance detection
Fecal DNA was isolated using the column‐based PureLink Microbiome DNA purification kit (cat: A29790, Thermo Fisher Scientific) according to the manufacturers' protocol. In short, 200 mg of fecal material was transferred to a tube with beads. Lysis buffer was added and lysis was facilitated through bead beating. The samples were loaded into the column together with a binding buffer and spun down at 14,000 g. The membrane of the column was subsequently washed and the DNA eluted.
DNA was then qualitatively and quantitatively tested using spectrophotometry before being used in the Viasure H. pylori + Clarithromycin resistance rtPCR detection kit (cat: VS‐CLA112L, CerTest Biotec) according to the manufacturers' protocol. In short, the wells were rehydrated before use, after which samples and controls were loaded. Plates were briefly centrifuged and loaded into a thermocycler (Thermo Fisher Scientific). The following program was used: polymerase activation 2 min at 95ºC followed by 45 cycles of denaturation for 10 s at 95ºC, annealing/extension for 50 s at 63ºC. Results were measured on the StepOnePlus Real‐Time PCR system (Thermo Fisher Scientific); the ROX channel was used for H. pylori, VIC for the wild type sequence of the 23S rRNA gene and the FAM channel for the clarithromycin resistance‐associated sequence of the 23S rRNA gene (to detect mutations A2142G and A2143G). Control sample provided by the manufacturer was used as a reference sample. Samples were assessed on three parameters: the Ct value has to be lower than 45, the amplification curve should have a sigmoidal shape and the fluorescence intensity should be larger than the negative control.
Statistical methods
The positive predictive value (PPV) comprised all participants diagnosed with H. pylori by the studied test proportionally to participants with a positive H. pylori UBT result multiplied by 100. The negative predictive value (NPV) comprised all participants with a negative H. pylori result by the studied test proportionally to participants with a negative H. pylori UBT result multiplied by 100. Sensitivity was calculated by dividing true positives by true positives plus false‐negative results, multiplied by 100. Specificity was calculated by dividing true negatives by true negatives plus false positives, multiplied by 100. Overall accuracy was calculated by dividing true positives and true negatives by all tests performed. Confidence intervals for sensitivity, specificity and accuracy are “exact” Clopper–Pearson confidence intervals, and confidence intervals for the predictive values are the standard logit confidence intervals. 19 Receiver operator characteristics (ROC) curves were plotted where the area under the curve (AUC) could be calculated. For interpretation an AUC >0.9 was considered “outstanding discrimination,” 0.8–0.9 was considered “excellent discrimination,” 0.7–0.8 as “acceptable discrimination,” 0.5–0.7 as “poor discrimination,” and 0.5 as “no discrimination.” 20 Differences between means were evaluated using a t test. A two‐sided significance level of p < 0.05 for all tests was used. Statistical analysis was performed using SPSS version 26.
RESULTS
Outcomes of H. pylori eradication
Baseline characteristics of the study population are shown in (Table 1) and have also been described previously. 18 Of the 182 patients in this cohort, 145 were documented to have been treated for H. pylori. Treatment of H. pylori was not part of the study protocol but was performed at the discretion of the treating physician as a part of the routine clinical care. The data were collected from electronic patient records and treatment regimens can therefore be regarded as “real world data” on H. pylori treatment in two large medical centers in the Netherlands. The majority of these patients received triple therapy, including PPI, clarithromycin, and amoxicillin (64.8%) or triple therapy, including PPI, amoxicillin, and levofloxacin (24.8%) as first‐line treatment. After a first round of H. pylori eradication treatment, with patients being prescribed antibiotics for a mean duration of 8.9 days (range 5–14 days), an eradication rate of 68.1% was achieved. After each subsequent round of eradication treatment, the eradication rate decreased, until it stabilized at around 25% after the third round of treatment. On average, a patient required 1.7 eradication treatments to achieve H. pylori eradication. All treatments and their outcomes are visualized in (Figure 2). Of the patients who were prescribed treatment for 7 days or less (68% of all patients), 63.9% achieved successful H. pylori eradication, which increased to 66.7% and 82.6% when patients were treated for 10 or 14 days, respectively (p = 0.07). In total, H. pylori eradication was eventually achieved in 130/145 patients (89.7%). Detailed information on eradication treatments may be found in the supplementary materials (Table S1).
TABLE 1.
Baseline statistics of the study cohort.
| Study population | 145 |
|---|---|
| Sex: Male n (%) | 62 (42.7%) |
| Age: Years (mean, SD) | 51.2 (SD 14.6) |
| Ethnicity: Western, n (%) | 82 (56.6%) |
| UBT: Helicobacter pylori positive n (%) | 51 (35.2%) |
| Total cured n (%) | 130 (89.7%) |
| Treatment duration: Days (mean, SD) | 8.86 (SD 2.66) |
Abbreviation: UBT, urea breath test.
FIGURE 2.
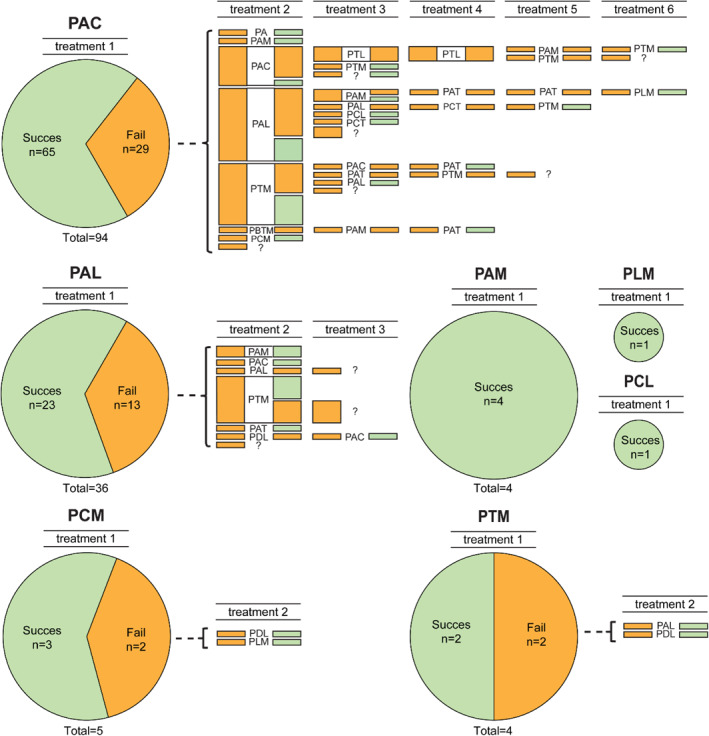
Visual representation of Helicobacter pylori eradication treatments received by study cohort and success rates thereof. PAC, PPI + Amoxicillin + Clarithromycin; PAL, PPI + Amoxicillin + Levofloxacin; PCM, PPI + clarithromycin + Metronidazole; PAM, PPI + Amoxicillin + Metronidazole; PLM, PPI + Levofloxacin + Metronidazole; PTM, PPI + Tetracyclin + Metronidazole; PCL, PPI + Clarithromycin + Levofloxacin; PBTM, PPI + Bismuth + Tetracyclin + Metronidazole; PA, PPI + Amoxicillin; PAT, PPI + Amoxicillin + Tetracyclin; PDL, PPI + Doxycyclin + Levofoxacin; PCT, PPI + Clarithromycin + Tetracyclin; PLM, PPI + Levofloxacin + Metronidazole; ?, unknown therapy.
Stool antigen measurements in dry stool and FIT are equally accurate to verify eradication of H. pylori
Guidelines advise either UBT or the less time‐consuming (fecal) SAT for verification of H. pylori eradication. We therefore investigated to what extent H. pylori antigen tests in fecal material collected by means of FIT may also be used to verify eradication in these patients. FIT and dry stool samples were available from 87 to 84 of the 88 patients undergoing UBT for verification of eradication, respectively (Figure 1). No difference in sensitivity (88.5% [95% CI 69.9–97.6] vs. 92.9% [95% CI 76.5–99.1]) or specificity (64.2% [95% CI 49.8–76.9] vs. 62.1% [95% CI 48.4–74.5]) was found between dry stool and FIT fluid when used for eradication verification, with the standard 3 ng/mL cutoff (Table 2). However, H. pylori antigen tests in both stool and FIT samples showed considerable false positives when compared with UBT, resulting in low specificity and PPV. The investigation of different cut‐off points to optimize test results demonstrated that using a higher cut‐off increases specificity and PPV, however at a cost for both sensitivity and NPV (Table 3). Receiver operator curves were plotted for stool and FIT eradication samples versus UBT results and showed comparable AUCs of 0.912 (95% CI 0.844–0.979, outstanding discrimination) and 0.837 (95% CI 0.741–0.932, excellent discrimination), respectively 20 (Figure S1).
TABLE 2.
Diagnostic performance of stool antigen tests in stool or FIT fluid for verification of Helicobacter pylori eradication, using urea breath test as the gold standard.
| Test | PR (%) (95% CI) | PPV (%) (95% CI) | NPV (%) (95% CI) | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | Accuracy (%) (95% CI) |
|---|---|---|---|---|---|---|
| FIT (n = 87) | 55.8 (44.7–66.5) | 62.1 (48.4–74.5) | 92.9 (76.5–99.1) | 94.7 (82.3–99.4) | 54.2 (39.2–68.6) | 72.1 (61.4–81.2) |
| Stool (n = 84) | 54.9 (43.5–65.9) | 64.2 (49.8–76.9) | 88.5 (69.9–97.6) | 91.9 (78.1–98.3) | 54.8 (38.7–70.2) | 72.2 (60.9–81.7) |
Abbreviations: CI, confidence interval; HP‐FIT, H. pylori antigen in fecal immunochemical test; NPV, negative predictive value; PPV, positive predictive value; PR, positivity rate.
TABLE 3.
Diagnostic performance of stool antigen tests in stool or FIT fluid for determination of verification of Helicobacter pylori eradication using different cutoffs.
| Cutoff | PPV (%) (95% CI) | NPV (%) (95% CI) | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | Accuracy (%) (95% CI) |
|---|---|---|---|---|---|
| FIT | |||||
| 3 ng | 62.1 (48.4–74.5) | 92.9 (76.5–99.1) | 94.7 (82.3–99.4) | 54.2 (39.2–68.6) | 72.1 (61.4–81.2) |
| 4 ng | 69.0 (55.5–80.5) | 82.1 (63.1–93.9) | 88.9 (76.0–96.3) | 56.1 (39.8–71.5) | 73.3 (62.2–82.2) |
| 5 ng | 75.9 (62.8–86.1) | 78.6 (59.1–91.7) | 88.0 (75.7–95.5) | 61.1 (43.5–76.9) | 76.7 (66.4–85.2) |
| 6 ng | 79.3 (66.7–88.8) | 71.4 (51.3–86.8) | 85.2 (72.9–93.4) | 62.5 (43.7–78.9) | 76.7 (66.4–85.2) |
| Stool | |||||
| 3 ng | 64.2 (49.8–76.9) | 88.5 (69.9–97.6) | 91.9 (78.1–98.3) | 54.8 (38.7–70.2) | 72.2 (60.9–81.7) |
| 4 ng | 78.6 (65.6–88.4) | 84.6 (65.1–95.6) | 91.7 (80.0–97.7) | 64.7 (46.5–80.3) | 80.5 (70.3–88.4) |
| 5 ng | 82.1 (69.6–91.1) | 80.8 (60.7–93.5) | 90.2 (78.6–96.7) | 67.7 (48.6–83.3) | 81.7 (71.6–89.4) |
| 6 ng | 89.5 (78.5–96.0) | 81.5 (61.9–93.7) | 91.1 (82.1–95.8) | 78.6 (62.8–88.9) | 86.9 (77.8–93.3) |
Abbreviations: CI, confidence interval; FIT, H. pylori antigen in fecal immunochemical test; NPV, negative predictive value; PPV, positive predictive value.
Overall, SAT in stool or FIT showed lower specificity for the detection of eradication than previously reported for H. pylori diagnosis. 18 We hypothesized that this might be due to trace amounts of H. pylori antigen still being present in the stool after eradication. However we found no significant differences between the quantitative values of H. pylori fecal antigen when we compared patients that tested negative for H. pylori and had no history of infection versus those patients that tested negative for H. pylori upon eradication treatment (3.0 [95% CI 2.0–4.0] versus 4.0 [95% CI 2.1–5.9] for stool, p = 0.07 and 4.0 [95%CI 2.3–5.7] versus 3.0 [95% CI 1.5–4.4] respectively for FIT, p = 0.4).
Clarithromycin resistance may be determined by rt‐PCR from DNA isolated from solid fecal material, but not FIT fluid
Next, we investigated the possibility of detecting H. pylori infection using rt‐PCR of microbial DNA isolated from fecal material. We used a sub‐selection of the complete cohort (including patients tested for H. pylori diagnosis in addition for those tested for eradication, n = 97), on the basis of availability of material (Table 1, Figure 1). As the used multiplex probe‐based assay is normally performed on DNA isolated from either H. pylori cultures or gastric biopsies, we first verified that it also generates reliable rt‐PCR curves on DNA isolated from fecal material (Figure S2a–d). While a significantly less intense fluorescence signal was observed compared to the supplied internal positive control, repeated measurements show good within‐sample correlations in 20 out of 21 tested samples, with 1 sample showing H. pylori positivity only in one of two replicates (for examples see Figure 2e,f). In contrast to DNA isolated from solid stool, FIT fluid samples did not consistently yield DNA of sufficient quantity or quality to produce reliable rt‐PCR data (Figure 2g,h). This is possibly due to much lower concentrations of fecal material in the collected FIT fluid, as a typical FIT sample contains approximately 10 mg of fecal material, while from a dry stool sample the advised 200 mg of fecal material may be used.
First, we compared the H. pylori rt‐PCR results of stool samples against the UBT of the whole cohort (Table 4). Rt‐PCR showed a sensitivity of 86.3% (95% CI 73.7–94.3) and a specificity of 87% (95% CI 73.7–95.1) for the detection of H. pylori. The analysis of the potential of the rt‐PCR test to specifically verify the eradication of H. pylori showed a sensitivity of 92.6% [95% CI 75.7–99.1] and specificity of 96.0% [95% CI 79.7–99.9] (Table 4).
TABLE 4.
Diagnostic performance of PCR tests in stool for the diagnosis of Helicobacter pylori (H. pylori) or the verification of H. pylori eradication.
| Test | PR (%) | PPV (%) (95% CI) | NPV (%) (95% CI) | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | Accuracy (%) (95% CI) |
|---|---|---|---|---|---|---|
| rt‐PCR (n = 97) | 52.3 | 87.9 (77.3–93.9) | 85.3 (74.2–92.1) | 86.3 (73.7–94.3) | 87.0 (73.7–95.1) | 86.6 (78.2–92.7) |
| rt‐PCR (eradication) (n = 52) | 50.0 | 96.2 (78.5–99.4) | 92.3 (75.9–97.9 | 92.6 (75.7–99.1) | 96.0 (79.7–99.9) | 94.2 (84.1–98.8) |
Abbreviations: CI, confidence interval; NPV, negative predictive value; PPV, positive predictive value; PR, positivity rate; rt‐PCR, real time polymerase chain reaction for H. pylori in fecal material.
Next, we investigated the clarithromycin resistance in our population by stool rt‐PCR. Of the 97 tested samples, 14 (14.4%) tested positive for the clarithromycin resistant variant of the 23S rRNA gene. One of these 14 patients was considered to be false positive for the rt‐PCR test, as the UBT was negative (the probe specific for H. pylori was also negative within the rt‐PCR). Of the remaining 13 patients (13.4%), nine were treated with clarithromycin‐containing antibiotic regimens, of which eight patients failed eradication treatment. One patient achieved eradication after 14 days of standard triple therapy, which may be explained by the presence of amoxicillin in this combination treatment—amoxicillin monotherapy is known to achieve reasonable success in H. pylori eradication in high clarithomycin‐resistant regions when proper acid reduction is achieved. 21 Three patients were treated by non‐clarithromycin containing regimens, and one received an unknown treatment.
H. pylori culture was performed due to failure of therapy for six of the patients (outside of the protocol for this study, on the preference of the treating physician) for whom we also investigated clarithromycin resistance through rt‐PCR. Rt‐PCR results were in 100% concordance with culture results from these six patients, with three cases of clarithromycin resistance and three cases of clarithromycin‐susceptible H. pylori.
DISCUSSION
Due to rising antibiotic resistance, H. pylori eradication remains a significant challenge in general clinical care. In this study, we report that the real world efficacy of H. pylori eradication in a cohort of patients in a low‐incidence region remains low, demonstrating that empirical eradication treatments have limited success rates. Verification of eradication may be tested by SAT in both dry stool and FIT samples with comparable accuracy. However, both lack specificity for H. pylori eradication testing when compared with UBT. However, we demonstrate that clarithromycin‐resistance testing in stool is feasible and accurate.
Confirmation of H. pylori eradication is essential for clinical decision making. We have previously shown that similar to H. pylori antigen testing in dry stool samples, testing in less‐invasive FIT‐fluids shows a sensitivity of 94.2% (95% CI 84.1–98.9) and a specificity of 69.7% (95% CI 60.2–78.1) in diagnosing active H. pylori infection. 18 In this follow‐up study we show that H. pylori antigen testing in traditional stool and FIT performs equally well for the verification of eradication of H. pylori. However, rt‐PCR showed lower sensitivity than antigen tests, potentially due to PCR inhibiting agents in feces. 22 , 23 All fecal‐based tests performed worse than UBT in particular with regard to specificity. While UBT measures the enzymatic activity of urease produced from viable bacteria, SAT and rt‐PCR detect bacterial remnants which may take some time to transit through the gastrointestinal tract. UBT for eradication verification is generally planned 2 weeks after the completion of eradication treatment, with fecal material collected before the UBT took place, perhaps not allowing sufficient transit times for bacterial remnants to be completely removed. Future studies using a longer time window of testing are needed to verify whether this may increase the specificity of the stool tests.
With encroaching antibiotic resistance rates, antibiotic susceptibility testing should be re‐evaluated and, if possible, implemented in clinical practice. In this study, we show the possibility of using DNA isolated from fecal material to determine H. pylori antibiotic resistance. The clarithromycin resistance rate in our cohort was 13.4%, which is in line with historical regional data (9.2%–18.1%). 7 , 8 While the assay used was designed for use on biopsies, our data holds promise for the use of non‐invasive methods for resistance testing. Of note, while DNA isolation from FIT was inefficient in this study, bacterial genome sequencing has been performed even for low abundance samples, and thus optimization of DNA isolation procedures may allow future use of FIT fluid as well. 24 It is also likely that antibiotic resistance for other antibiotics may be investigated using the same approach, further guiding to a more personalized antibiotic treatment. 25 , 26 , 27
In our study, a significant proportion of tested samples for antibiotic resistance were tested to verify eradication. However, knowing the resistance state at initial diagnosis (i.e. prior to first treatment) would be even more useful. Future studies will have to test the cost efficacy and health benefits of such a test for early detection of H. pylori resistance. Limitations of the rt‐PCR test for determining H pylori presence and resistance (i.e. lower specificity than UBT and increased costs) could be offset by its use as an add‐on test to the SAT. In this scenario, a positive SAT would be followed by an rt‐PCR on the leftover material to determine H. pylori resistance, guiding the physician in prescribing their treatment (Figure 3). Performing rt‐PCR analysis only in the ∼25% of positive patients reduces the costs, while also negating the slightly lower sensitivity of the PCR‐based test when compared to the SAT. Given the inferior specificity of both stool and FIT in the eradication tests, the UBT remains the preferred test to verify eradication. This diagnostic scheme may contribute to a better use of antibiotics and hopefully reduce microbial antibiotic resistance. Further research into the efficacy as well as cost effectiveness of this approach is warranted.
FIGURE 3.

A diagnostic and treatment scheme incorporating a possible place for the fecal antibiotic susceptibility PCR. PCR, polymerase chain reaction; SAT, stool antigen test; UBT, urease breath test; standard triple therapy, PPI + clarithromycin + amoxicillin for 10–14 days; Bismuth containing quadruple therapy, PPI + bismuth + tetracyclin + metronidazole for 14 days.
Our data suggest that even within two centers within the same region in a small country like the Netherlands there is large heterogeneity in the treatment of patients for H. pylori, with 14 different antibiotic regimens being prescribed. Of note, our study did not guide the clinician treatment of H. pylori and the collected data therefore reflect real world data. The failure of eradication is likely a combination of rising antibiotic resistance of H. pylori as well as imperfect patient compliance and physician compliance to current guidelines. 5 , 6 , 8 The eradication rate in this cohort is lower than generally considered acceptable, as antimicrobial stewardship standards for empirical therapies are deemed to be around 90%. 7 , 28 Such high eradication rates are rarely achieved with current H. pylori treatment regimens, as in the Hp‐EuReg study standard triple therapy was only successful in 68% of cases while all treatment regimens combined averaged at a cure rate of 73.5% in the intention to treat analyses. 7 The application of systematic and evidence‐based approaches to prescription decision making may lead to better outcomes. 29 For example, the repetition of the same regimen after initial failure resulted in only one eradication in 11 attempts in our cohort. As a second example, only 22.7% of our patients received first‐line eradication treatment in concordance with regional and international standards. 5 Similar to others, our study indicates that a first treatment of 14 days (as recommended by guidelines) results in cure rates of 82.6% compared to 63.9% for a 7 days treatment, and having to resort to additional treatments incrementally reduces the cure rate. 7 , 28 , 30 Nevertheless, 7‐day regimes are still prescribed in 68% of cases in daily practice. Such inefficient use of drug treatments is likely to contribute to rising microbial antibiotic resistance worldwide. A possible reason for the high percentage of patients who were treated too shortly is the fact that Dutch guidelines for general practitioners do not comply with both national and international treatment guidelines and still advise 7 days treatment with triple therapy. The findings of this study underline the importance of elongating the eradication treatment. 5 , 31 , 32
We acknowledge several limitations to our study. First, as we did not obtain gastric biopsies, we had no opportunity to confirm the rt‐PCR results with the gold standard of culture for H. pylori presence and the detection of clarithromycin resistance on biopsies as is the gold standard for the rt‐PCR kit and the gold standard for the determination of antibiotic resistance. However, our resistance rates are in line with literature findings, and treatment and H. pylori culture histories which could be retrieved were in concordance with resistance rt‐PCR data. Second, golden standards for H. pylori diagnosis may be difficult to define, as the presence of other urease‐producing bacteria which are known to inhabit the oral cavity and stomach in some individuals may also cause false positives for the UBT. 33 However, this would have resulted in an underestimation of the performance of stool‐based antigen tests and rt‐PCR.
CONCLUSION
This study shows that H. pylori antigen testing in stool or FIT achieves equal accuracy; however, it seems inferior to UBT in assessing eradication success. Nevertheless, fecal samples may be used to non‐invasively determine antibiotic resistance by performing rt‐PCR after a positive SAT. Further research should be performed to confirm whether this approach is cost effective and will result in fewer prescriptions of ineffective antibiotics. Moreover, resistance to other antibiotics may be evaluated using additional probes in the rt‐PCR setup.
AUTHOR CONTRIBUTIONS
Michiel C. Mommersteeg, Stella A. V. Nieuwenburg, Manon C. W. Spaander and Gwenny M. Fuhler had full access to all study data and are responsible for data integrity and accuracy of the data analysis. Study concept and design: Michiel C. Mommersteeg, Stella A. V. Nieuwenburg, Manon C. W. Spaander, Maikel P. Peppelenbosch, Gwenny M. Fuhler. Acquisition of data: Michiel C. Mommersteeg, Stella A. V. Nieuwenburg, Leonieke M. M. Wolters, and Buddy H. C. M. Roovers. Analysis and interpretation of data: all coauthors. Drafting of the manuscript: Michiel C. Mommersteeg, Stella A. V. Nieuwenburg and Gwenny M. Fuhler. Critical revision of the manuscript for important intellectual content: all authors. Statistical analysis: Michiel C. Mommersteeg, Stella A. V. Nieuwenburg Obtained funding: Manon C. W. Spaander, Stella A. V. Nieuwenburg, Gwenny M. Fuhler. Administrative, technical, or material support: Auke P. Verhaar, Study supervision: Manon C. W. Spaander, Gwenny M. Fuhler and Maikel P. Peppelenbosch.
CONFLICT OF INTEREST STATEMENT
MCM declares that he has no conflicts of interest. SAVN declares that she has no conflict of interest. LMMW declares that she has no conflict of interest. BHCMR declares that she has no conflict of interest. AJV declares that she has no conflicts of interest. APV declares that she has no conflict of interest. MPP declares that he has no conflicts of interest. GMF declares that she has no conflict of interest. MJB has received research grants from Boston Scientific, grants and personal fees from Cook Medical, grants from Pentax Medical, grants from 3M, grants from Mylan, and grants from InterScope, outside the submitted work. EJK declares that he has no conflict of interest. MCWS has received grants from sysmex, sentinel, Medtronic, Boston Scientific, Norgine, outside the submitted work. The authors have no potential conflicts of interest to disclose that are relevant to this manuscript. Full disclosures have been submitted to the journal.
Supporting information
Supporting Information S1
Table S1
ACKNOWLEDGMENTS
The authors would like to acknowledge J. Francke, M. Ouwendijk and F. van Deurzen for their assistance in the execution of the UBT tests in the Erasmus MC. We would also like to acknowledge A. van Liere‐Baron for her assistance with organizational tasks and logistics at the Albert Schweitzer Hospital. This study was supported by the Dutch Digestive Foundation (D18‐02).
Mommersteeg MC, Nieuwenburg SAV, Wolters LMM, Roovers BHCM, van Vuuren HAJ, Verhaar AP, et al. The use of non‐invasive stool tests for verification of Helicobacter pylori eradication and clarithromycin resistance. United European Gastroenterol J. 2023;11(9):894–903. 10.1002/ueg2.12473
Michiel C. Mommersteeg and Stella A. V. Nieuwenburg share first authorship.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Correa P, Piazuelo MB. The gastric precancerous cascade. J Dig Dis. 2012;13(1):2–9. 10.1111/j.1751-2980.2011.00550.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liou J‐M, Malfertheiner P, Lee Y‐C, Sheu B‐S, Sugano K, Cheng H‐C, et al. Screening and eradication of Helicobacter pylori for gastric cancer prevention: the Taipei global consensus. Gut. 2020;69(12):2093–2112. 10.1136/gutjnl-2020-322368 [DOI] [PubMed] [Google Scholar]
- 3. European Health Union . A new EU approach on cancer detection – screening more and screening better. European commision; 2022. [press release] https://ec.europa.eu/ [Google Scholar]
- 4. Bujanda L, Nyssen OP, Vaira D, Saracino IM, Fiorini G, Lerang F, et al. Antibiotic resistance prevalence and trends in patients infected with Helicobacter pylori in the period 2013‐2020: results of the European Registry on H. pylori management (Hp‐EuReg). Antibiotics. 2021;10(9):1058. 10.3390/antibiotics10091058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Malfertheiner P, Megraud F, Rokkas T, Gisbert JP, Liou JM, Schulz C, et al. Management of Helicobacter pylori infection: the Maastricht VI/Florence consensus report. Gut. 2022;71(9):1724–1762. 10.1136/gutjnl-2022-327745 [DOI] [Google Scholar]
- 6. Megraud F, Bruyndonckx R, Coenen S, Wittkop L, Huang TD, Hoebeke M, et al. Helicobacter pylori resistance to antibiotics in Europe in 2018 and its relationship to antibiotic consumption in the community. Gut. 2021;70(10):1815–1822. 10.1136/gutjnl-2021-324032 [DOI] [PubMed] [Google Scholar]
- 7. Nyssen OP, Bordin D, Tepes B, Perez‐Aisa A, Vaira D, Caldas M, et al. European Registry on Helicobacter pylori management (Hp‐EuReg): patterns and trends in first‐line empirical eradication prescription and outcomes of 5 years and 21 533 patients. Gut. 2021;70(1):40–54. 10.1136/gutjnl-2020-321372 [DOI] [PubMed] [Google Scholar]
- 8. Ruiter R, Wunderink HF, Veenendaal RA, Visser LG, de Boer MGJ. Helicobacter pylori resistance in the Netherlands: a growing problem? Neth J Med. 2017;75(9):394–398. [PubMed] [Google Scholar]
- 9. Sugimoto M, Furuta T. Efficacy of tailored Helicobacter pylori eradication therapy based on antibiotic susceptibility and CYP2C19 genotype. World J Gastroenterol. 2014;20(21):6400–6411. 10.3748/wjg.v20.i21.6400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wenzhen Y, Yumin L, Quanlin G, Kehu Y, Lei J, Donghai W, et al. Is antimicrobial susceptibility testing necessary before first‐line treatment for Helicobacter pylori infection? Meta‐analysis of randomized controlled trials. Intern Med. 2010;49(12):1103–1109. 10.2169/internalmedicine.49.3031 [DOI] [PubMed] [Google Scholar]
- 11. Wilson DN. Ribosome‐targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014;12(1):35–48. 10.1038/nrmicro3155 [DOI] [PubMed] [Google Scholar]
- 12. Vester B, Douthwaite S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother. 2001;45(1):1–12. 10.1128/aac.45.1.1-12.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pichon M, Pichard B, Barrioz T, Plouzeau C, Croquet V, Fotsing G, et al. Diagnostic accuracy of a noninvasive test for detection of Helicobacter pylori and resistance to clarithromycin in stool by the amplidiag H. pylori+ClariR real‐time PCR assay. J Clin Microbiol. 2020;58(4). 10.1128/jcm.01787-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Redondo JJ, Keller PM, Zbinden R, Wagner K. A novel RT‐PCR for the detection of Helicobacter pylori and identification of clarithromycin resistance mediated by mutations in the 23S rRNA gene. Diagn Microbiol Infect Dis. 2018;90(1):1–6. 10.1016/j.diagmicrobio.2017.09.014 [DOI] [PubMed] [Google Scholar]
- 15. Marrero Rolon R, Cunningham SA, Mandrekar JN, Polo ET, Patel R. Clinical evaluation of a real‐time PCR assay for simultaneous detection of Helicobacter pylori and genotypic markers of clarithromycin resistance directly from stool. J Clin Microbiol. 2021;59(5). 10.1128/jcm.03040-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun L, Talarico S, Yao L, He L, Self S, You Y, et al. Droplet digital PCR‐based detection of clarithromycin resistance in Helicobacter pylori isolates reveals frequent heteroresistance. J Clin Microbiol. 2018;56(9). 10.1128/jcm.00019-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Beckman E, Saracino I, Fiorini G, Clark C, Slepnev V, Patel D, et al. A novel stool PCR test for Helicobacter pylori may predict clarithromycin resistance and eradication of infection at a high rate. J Clin Microbiol. 2017;55(8):2400–2405. 10.1128/jcm.00506-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nieuwenburg SAV, Mommersteeg MC, Wolters LMM, van Vuuren AJ, Erler N, Peppelenbosch MP, et al. Accuracy of H. pylori fecal antigen test using fecal immunochemical test (FIT). Gastric Cancer. 2022;25(2):375–381. 10.1007/s10120-021-01264-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mercaldo ND, Lau KF, Zhou XH. Confidence intervals for predictive values with an emphasis to case‐control studies. Stat Med. 2007;26(10):2170–2183. 10.1002/sim.2677 [DOI] [PubMed] [Google Scholar]
- 20. Hosmer DW, Lemeshow S, Sturdivant RX. Applied logistic regression. Hoboken, NJ: Wiley; 2013. Available from: Cover image http://catalogimages.wiley.com/images/db/jimages/9780470582473.jpg [Google Scholar]
- 21. Furuta T, Yamade M, Kagami T, Uotani T, Suzuki T, Higuchi T, et al. Dual therapy with vonoprazan and amoxicillin is as effective as triple therapy with vonoprazan, amoxicillin and clarithromycin for eradication of Helicobacter pylori . Digestion. 2020;101(6):743–751. 10.1159/000502287 [DOI] [PubMed] [Google Scholar]
- 22. Thornton CG, Passen S. Inhibition of PCR amplification by phytic acid, and treatment of bovine fecal specimens with phytase to reduce inhibition. J Microbiol Methods. 2004;59(1):43–52. 10.1016/j.mimet.2004.06.001 [DOI] [PubMed] [Google Scholar]
- 23. Acharya KR, Dhand NK, Whittington RJ, Plain KM. PCR inhibition of a quantitative PCR for detection of Mycobacterium avium subspecies paratuberculosis DNA in feces: diagnostic implications and potential solutions. Front Microbiol. 2017;8:115. 10.3389/fmicb.2017.00115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grobbee EJ, Lam SY, Fuhler GM, Blakaj B, Konstantinov SR, Bruno MJ, et al. First steps towards combining faecal immunochemical testing with the gut microbiome in colorectal cancer screening. United Eur Gastroenterol J. 2020;8(3):293–302. 10.1177/2050640619890732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cambau E, Allerheiligen V, Coulon C, Corbel C, Lascols C, Deforges L , et al. Evaluation of a new test, genotype HelicoDR, for molecular detection of antibiotic resistance in Helicobacter pylori . J Clin Microbiol. 2009;47(11):3600–3607. 10.1128/jcm.00744-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Contreras M, Benejat L, Mujica H, Pena J, Garcia‐Amado MA, Michelangeli F, et al. Real‐time PCR detection of a 16S rRNA single mutation of Helicobacter pylori isolates associated with reduced susceptibility and resistance to tetracycline in the gastroesophageal mucosa of individual hosts. J Med Microbiol. 2019;68(9):1287–1291. 10.1099/jmm.0.001051 [DOI] [PubMed] [Google Scholar]
- 27. Correa P, Piazuelo MB, Wilson KT. Pathology of gastric intestinal metaplasia: clinical implications. Am J Gastroenterol. 2010;105(3):493–498. 10.1038/ajg.2009.728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Graham DY. Transitioning of Helicobacter pylori therapy from trial and error to antimicrobial stewardship. Antibiotics. 2020;9(10):671. 10.3390/antibiotics9100671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nyssen OP, Vaira D, Tepes B, Kupcinskas L, Bordin D, Perez‐Aisa A, et al. Room for improvement in the treatment of Helicobacter pylori infection: lessons from the European Registry on H. pylori management (Hp‐EuReg). J Clin Gastroenterol. 2022;56(2):e98–e108. 10.1097/mcg.0000000000001482 [DOI] [PubMed] [Google Scholar]
- 30. Graham DY. Hp‐normogram (normo‐graham) for assessing the outcome of H. pylori therapy: effect of resistance, duration, and CYP2C19 genotype. Helicobacter. 2016;21(2):85–90. 10.1111/hel.12287 [DOI] [PubMed] [Google Scholar]
- 31. SWAB . SWAB richtlijn Helicobacter, Pylori: SWABID; 2023. [antibiotic guideline]. https://adult.nl.antibiotica.app/en/node/8049
- 32. De Jongh EDWN, Numans ME, Smeink P, Van der Weele GM, Wesseler GH. Beleid na positieve H. pylori test richtlijne. nhg.org: NHG; 2021. [General practioner guideline]. https://richtlijnen.nhg.org/standaarden/maagklachten#samenvatting‐beleid‐na‐positieve‐h‐pylori‐test
- 33. Furlong ST, Caulfield JP. Head group precursors modify phospholipid synthesis in Schistosoma mansoni. J Lipid Res. 1991;32(4):703–712. 10.1016/s0022-2275(20)42058-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information S1
Table S1
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


