Figure 1.
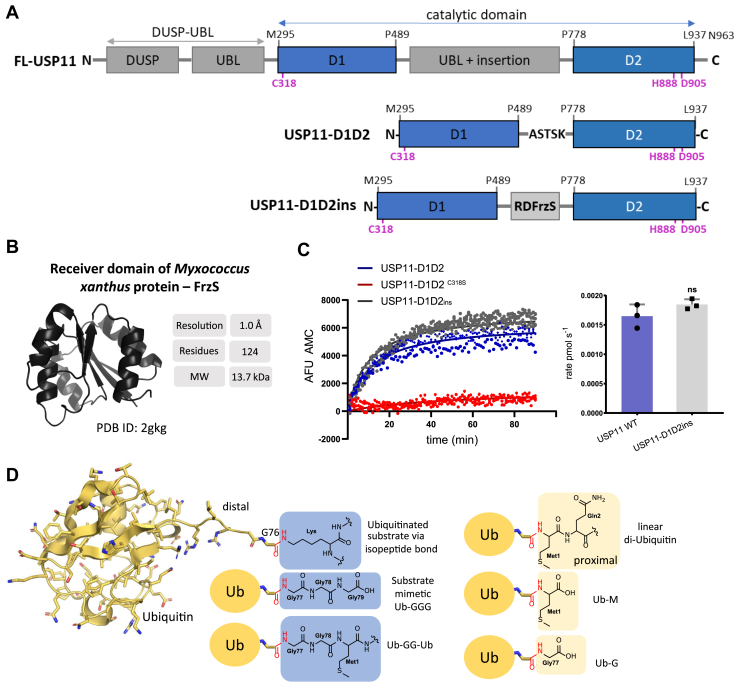
Design of USP11 catalytic domain and ubiquitin substrate constructs. A, top: schematic representation of the hUSP11 domain structure (UniProt P51784). The catalytic subdomains D1 and D2 are depicted in blue and the catalytic triad residues marked in magenta. Additional domains are shown in gray. Below: schematic representation of the USP11-D1D2 (residues Met295-Leu937) construct with the insertion replaced by an ASTSK linker or the RDFrzS loop insertion tag shown in light gray to engineer USP11-D1D2ins. B, receiver domain structure from the Myxococcus xanthus protein FrzS (26) used as an insertion tag. C, progress curves of USP11-D1D2 ubiquitin-7-amino-4-methylcoumarin cleavage in comparison with USP11-D1D2 C318S and USP11-D1D2ins constructs with a bar chart (unpaired two-tailed t test; n = 3 independent experiments; error = SD) highlighting that the insertion does not significantly affect the catalytic activity. D, ubiquitin C-terminal tail modified substrates as used in this study with respective tail modifications shown as chemical structures. USP, ubiquitin-specific protease.