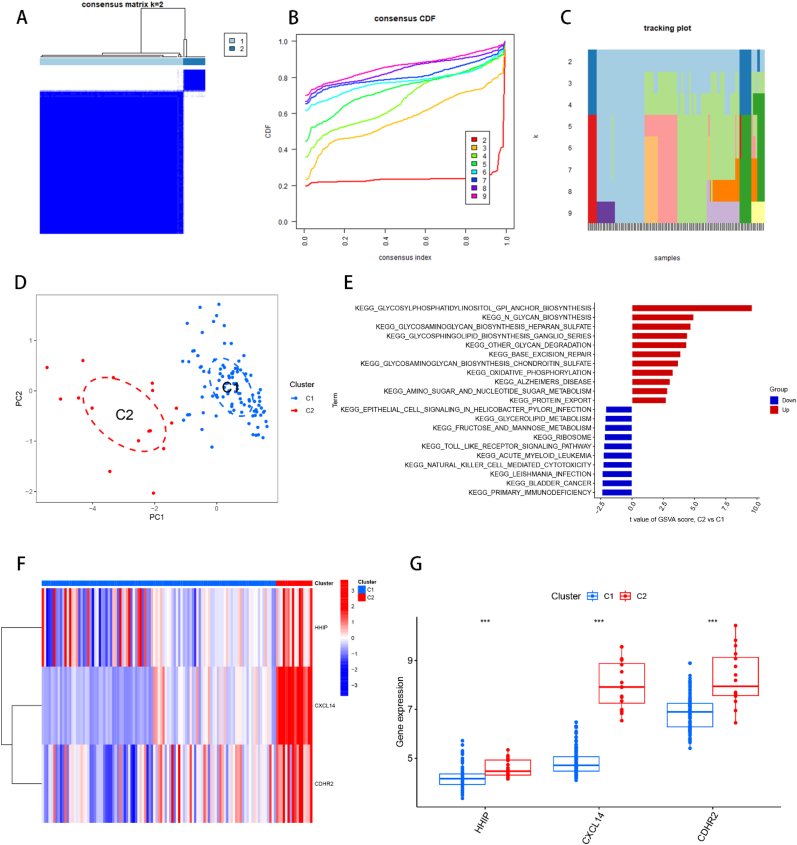
Fig. 10.
Consensus clustering of HBV-HCC samples. (A) Consensus matrix plot, the cleaner the blank area between the blue modules indicates the more successful analysis. (B) Cumulative distribution function (CDF) plot of consensus clustering, showing the relative change of consensus index from k = 2 to k = 9 with the change of CDF value, and the k value of the curve with the most stable change is the optimal fractal number. (C) Trace plot of k = 2 to k = 9. (D) PCA plot of HBV-HCC samples. The scatter plot allows visualization of the characteristic genes that classify HBV-HCC into two subtypes, C1 and C2. (E) GSVA analysis between C1 and C2, red represents the up-regulated functional pathway and blue represents the down-regulated functional pathway. (F) Heat map between C1 and C2 and feature genes, red indicates up-regulated gene expression and blue indicates down-regulated gene expression. (G) Box line plot of the difference in expression of feature genes between C1 and C2 of two subtypes of HBV-HCC. *p < 0.05, **p < 0.01, ***p < 0.001.