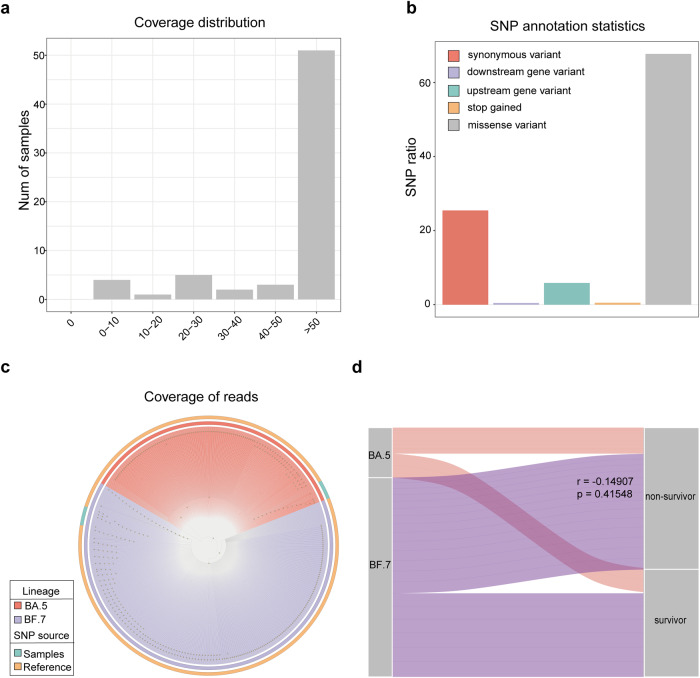
Fig. 1.
Analysis of SARS-CoV-2 genomes in the BALF samples. a Coverage distribution of reads mapped to reference SARS-CoV-2 genome, in which we found 8 complete genomes and 43 > 50% completeness. COVID-19 patients (n = 63). b Overview of SNP variant types in the assembled genome, in total we found 34 insertions, 18 deletions, and 583 substitutions. COVID-19 patients (n = 51). c Phylogenetic relationships of the 51 genomes assembled in this study vs. 682 SARS-CoV-2 genomes sampled in Beijing general population from the same period and deposited in GISAID. 20 genomes belong to BA.5 and 24 to BF.7 lineages. COVID-19 patients (n = 44). d The correlation between SARS-CoV-2 genomes of BF.7 and BA.5 lineages and 28-day mortality (survivors or non-survivors) based on Spearman’s rank test, no significant differences were found between the 28-day mortality rate in the two strains. COVID-19 patients (n = 32)