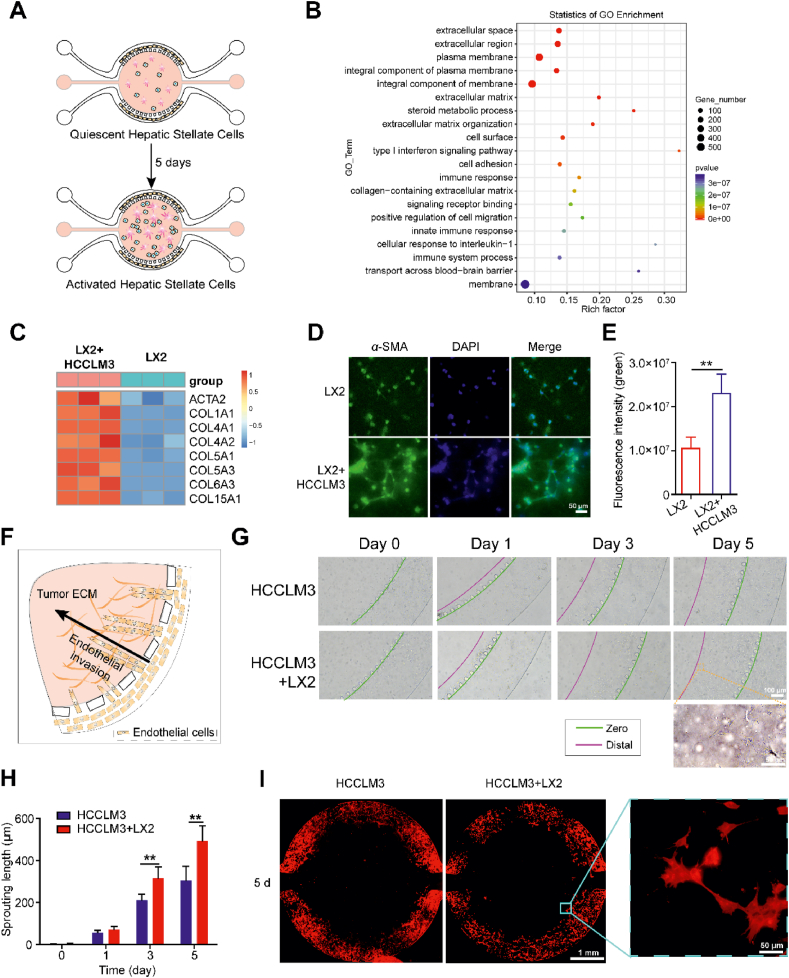
Figure 2.
Activated HSCs contributed to endothelial cell invasion. (A) Scheme of hepatic stellate cell activation in the HCC-on-a-chip. LX2 cells (no label) and tumor cells (labeled with CFDA-SE, green) were mixed with the Matrigel at a 1:1 ratio (total density 10 × 106 cells/mL) and injected in the HCC-on-a-chip for 5 days. After 5 days' culture, HCCLM3 cells were selectively removed which were labeled in green fluorescence forehand for the isolation of pure LX2 cells by flow cytometry cell sorting. (B) The RNA-seq was performed to analyze the total RNA from the isolated LX2 cells via chip (n = 3 per group). Gene Ontology enrichment analysis was applied to show the upregulated genes and enriched biological processes in LX2 cells from the cell co-culture group. (C) Heat map of multiple genes associated with HSCs activation were shown in the indicated groups (n = 3 per group). (D) Representative image showing α-SMA immunofluorescence staining of LX2 cells in the HCC-on-a-chip model (n = 3 per group). Scale bar: 50 μm. (E) Quantification of the α-SMA fluorescence intensity in indicated groups. (F) Schematic representation of HUVEC in the lateral channels invading towards the central chamber in response to the tumor cytokines microenvironments. (G) Representative bright-field images of endothelial cell sprout in the microfluidic device were shown on Days 1, 3 and 5. (H) Quantification of endothelial cell invasion length at determined time points in the indicated groups (n = 3 per group). (I) Representative images of endothelial cell sprout after F-actin staining. Scale bar: 1 mm. Data are presented as mean ± SEM. ∗P < 0.05; ∗∗P < 0.01.