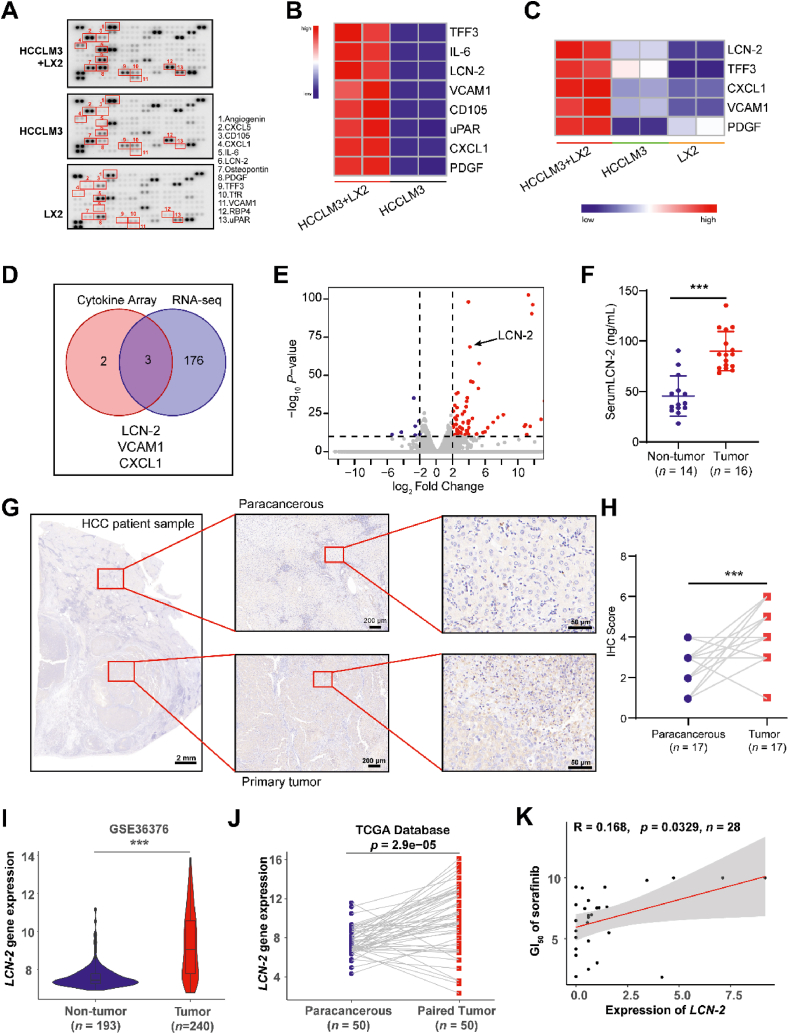
Figure 5.
LCN-2 was identified as a key factor in HCC development. (A) The HCCLM3 cells and LX2 were cultured in the chip for 5 days and the conditional medium was gathered from the channel for cytokine array assessment. Each cell line cultured alone was set as control. Cytokines were indicated in red which showed a difference in secretion. (B) Heat map analysis showed the significant changed cytokines in the cell co-culture group compared with HCCLM3 cells culture alone. (C) Heat map analysis showed the significant changed cytokines in the cell co-culture group compared with each cell line culture alone. (D) The total RNA of isolated HCCLM3 cells from the HCC-on-a-chip was applied for RNA-seq assessment. Venn diagram indicated the overlapped genes with a dramatic increase by combining the results of RNA-seq in HCCLM3 cells and cytokines analysis in C. (E) The Volcano plot analysis of RNA-seq data in isolated HCCLM3 cells from chip. Three upregulated cytokines were marked which showed an obvious increase of secretion level in the cell co-culture system via chip. (F) LCN-2 expression levels in the serum of non-tumor (n = 14) and HCC patients (n = 16). (G) LCN-2 expression was detected by IHC in paracancerous and primary HCC tumor samples. Scale bar: 2 mm. (H) IHC score analysis of LCN-2 expression across paracancerous and primary tumor sites (n = 17 per group). (I) LCN-2 gene expression analysis in GSE36376 datasets from GEO database containing 193 non-tumor samples and 240 HCC tumor samples. (J) LCN-2 gene expression analysis in 50 paired normal and HCC samples from TCGA database. (K) Correlation between LCN-2 expression and sorafenib GI50, based on the liver cancer cell lines (n = 28) from CTRP. The linear relationship was determined by a two-tailed Pearson correlation analysis. ∗P < 0.05; ∗∗P < 0.01.