Table 1.
List of representative anticancer small molecules by targeting E2s, CRLs or DUBs.
| Name | Structure | Target | In vitro | In cells | Cell viability, IC50 | Cancer cell lines | Ref. |
|---|---|---|---|---|---|---|---|
| CC0651 | 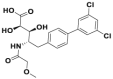 |
Cdc34 | IC50 (FP) = 2.5 μmol/L | p27 deubiquitination @ 30 μmol/L | HCT116 | 280,281 | |
| Leucettamol A | 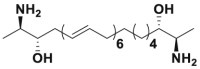 |
Ubc13 | 105 μmol/L | 282 | |||
| 2D-08 | 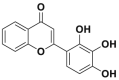 |
Ubc9 | IC50 = 6 μmol/L | 100 μmol/L | BT-474 | 283 | |
| Compound 25 | 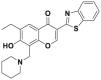 |
CRL1Skp2 | >80% inhibition@5 μmol/L | p21, p27 deubiquitination @ 5 μmol/L | <10 μmol/L | PC-3, LNCaP | 293 |
| C1, C2, C16, C20 | 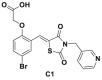 |
>80% inhibition@10 μmol/L | p21, p27 deubiquitination @ 10 μmol/L | MCF-7, TD47, LNCaP | 294 | ||
| VH298 | 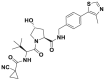 |
CRL2VHL | IC50 (FP) = 80 nmol/L; Kd (ITC) = 90 nmol/L |
HIF-1α destabilization @ 50 μmol/L | No cytotoxic | Hela | 299 |
| CM11 | homo-PROTAC | Kd (ITC) = 25 nmol/L | HIF-1α destabilization @ 1 μmol/L | Hela | 300 | ||
| CDDO-Me | 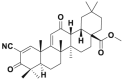 |
CRL3Keap1 | 58.9%@0.1 μmol/L | 50–160 nmol/L | MCF-7 | 302 | |
| ML334 | 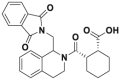 |
Kd = ∼ 1 μmol/L | EC50 = 18 μmol/L | U2OS | 303 | ||
| Compound 2 | 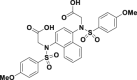 |
EC50 = 28.6 nmol/L | Nrf2–ARE induction@10 μmol/L | HepG2-ARE-C8 | 304 | ||
| KI696 | 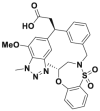 |
Kd (ITC) = 1.3 nmol/L | 305 | ||||
| 6lc | 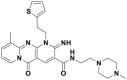 |
CRL3SPOP | KD = 30 μmol/L | PTEN deubiquitination @ 15 μmol/L | 2.1–3.5 μmol/L | A498, OS-RC-2 | 308,309 |
| CC885 | 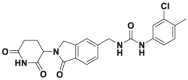 |
CRL4CRBN | GSPT1 degradation @ 1 nmol/L | NB4 | 189 | ||
| Thalidomid | 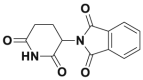 |
336 | |||||
| Pomalidomide | 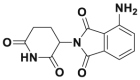 |
337 | |||||
| Lenalidomid | 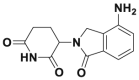 |
IKZF1 and IKZF3 degradation @ 1 μmol/L | MM1S | 191,338 | |||
| Indisulam |  |
CRL4DCAF15 | RBM39 degradation @ 2 μmol/L | HCT116 | 339 | ||
| CR8 | 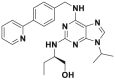 |
Cyclin K degradation @ 1 μmol/L | HCT116 | 340 | |||
| HQ461 | 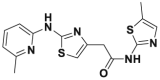 |
DC50 = 0.13 μmol/L | 1.5 μmol/L | A549 | 341 | ||
| dCeMM2/3/4 | 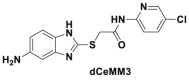 |
Cyclin K degradation @ 2.5 μmol/L | KBM7 | 342 | |||
| SJB2-043 | 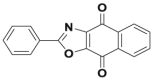 |
USP1 | 0.54 μmol/L | >80% inhibition @ 1 μmol/L | 1.1 μmol/L | K562 | 347 |
| ML323 | 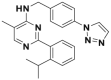 |
76 nmol/L | PCNA deubiquitination @ 20 μmol/L | 3 μmol/L | H1299 | 348 | |
| Q29 | 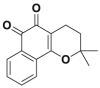 |
USP2 | 82% inhibition @ 0.5 μmol/L | NA | 4.7 μmol/L | DU145 | 350 |
| ML364 | 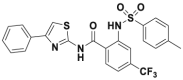 |
1.1–1.7 μmol/L | cyclin D1 destabilization @ 0.97 μmol/L | 3 μmol/L | HCT116 | 351 | |
| LCAHA | 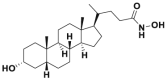 |
9.7 μmol/L | cyclin D1 destabilization @ 20 μmol/L | 0.9 μmol/L | HCT116 | 352 | |
| XL188 | 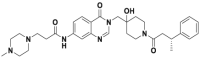 |
USP7 | 90 nmol/L | p53 accumulation @5 μmol/L | MM.1S, MCF7 | 357 | |
| FT671 | 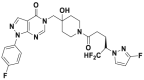 |
52 nmol/L | p53 accumulation @10 μmol/L | 33 nmol/L | HCT116, MM.1S, MCF7 | 358 | |
| Compound 4 | 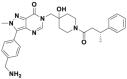 |
6 nmol/L | p53 accumulation @1 μmol/L | 2–29 nmol/L | HCT116 | 359 | |
| Compound 41 | 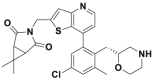 |
0.44 nmol/L | p53 accumulation @25 nmol/L | 0.09–0.45 μmol/L | MM.1S, H526 | 360 | |
| GNE6640 | 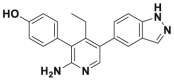 |
0.75 μmol/L | p53 accumulation @10 μmol/L | >50% inhibition@5 μmol/L | HCT116, MCF7, EOL-1 | 361 | |
| Capzimin | 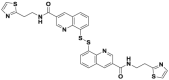 |
Rpn11 | 0.39 μmol/L | Deubiquitination @ 2–10 μmol/L | 2.1–3.8 μmol/L | 293T, A549, HCT116 | 364 |
| SOP11 | 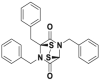 |
1.3/0.6 μmol/L | Deubiquitination @ 10 μmol/L | GI50 = 4.7 μmol/L | HCT116 | 365 | |
| VLX1570 | 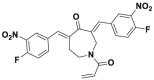 |
USP14/UCH37 | 8.1/14 μmol/L | >80% inhibition @ 25 mmol/L | 43–126 nmol/L | RPMI8226, KMS11, OPM-2 | 366 |
