Figure 1.
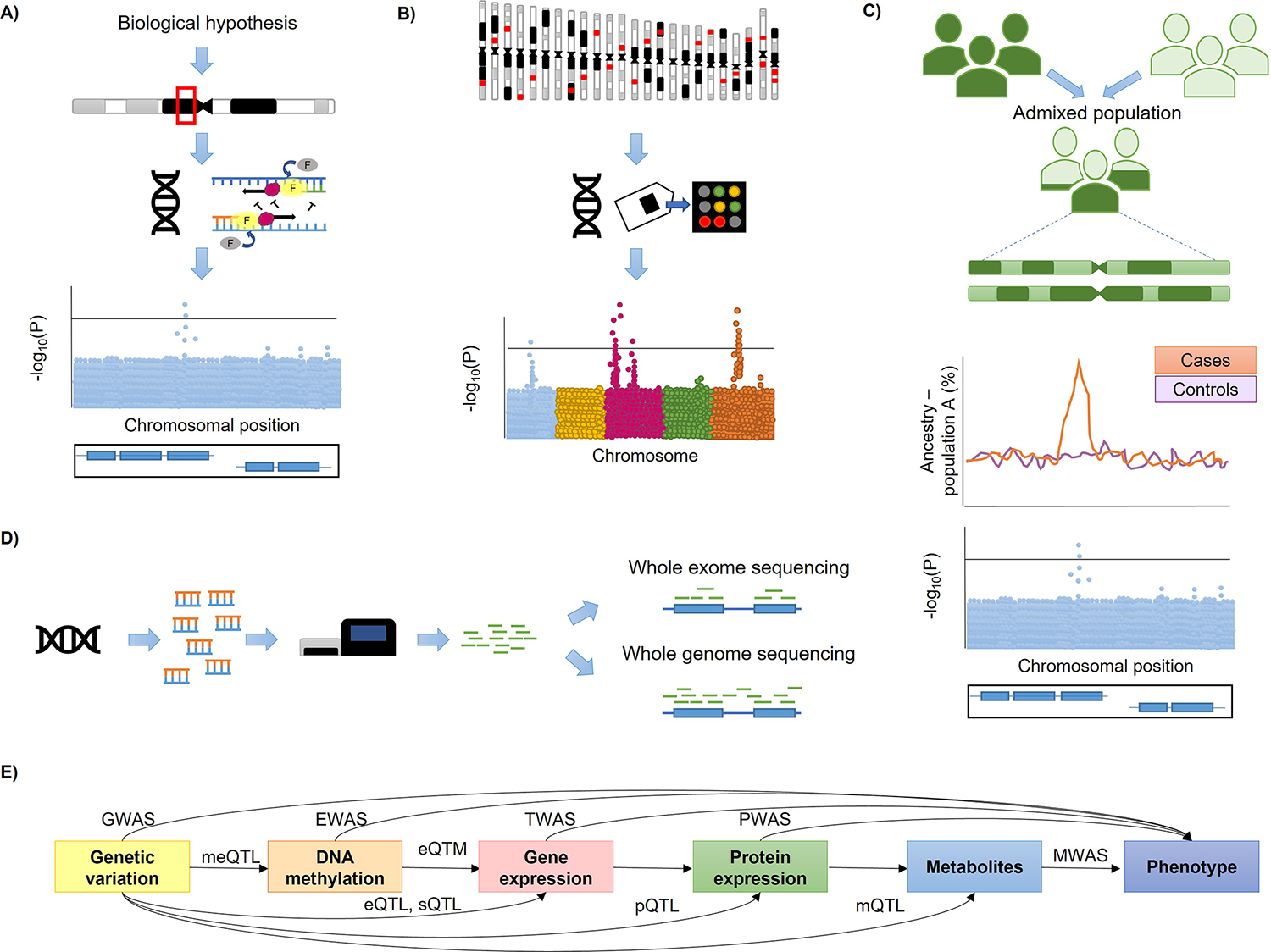
A) Candidate-gene association study. A biological hypothesis is used to prioritize genomic regions that will be genotyped, and genetic variants within the regions will be tested for association with the trait of interest. B) GWAS. Genetic variation is profiled via genome-wide genotyping arrays and evaluated for association with the trait of interest. C) Theoretical framework of admixture mapping. The genomes of the admixed individuals are composed of mosaics of ancestral blocks derived from ancestral populations. The association of local ancestry and a trait of interest is evaluated in order to prioritize genomic regions where genetic variants will be assessed for association with the trait of interest. D) Next-generation sequencing (NGS) approaches. The DNA is fragmented and sequenced, then reads are mapped to the reference genome. While whole exome sequencing (WES) focuses on genomic protein-coding regions (exons), whole genome sequencing (WGS) determines genetic variation in any part of the genome. E) Combination of different -omic and clinical layers to understand the biological mechanisms underlying a trait of interest. The association of genetic variation with a specific trait, DNAm, gene expression, protein expression or metabolites is assessed by GWAS, epigenome-wide association studies (EWAS), transcriptome-wide association studies (TWAS), proteome-wide association studies (PWAS) or metabolome-wide association studies (MWAS), respectively. Moreover, a regulatory genetic variant can exert effects as methylation quantitative trait locus (meQTL), expression quantitative trait locus (eQTL), splicing quantitative trait locus (sQTL), protein quantitative trait locus (pQTL) and/or metabolic quantitative trait locus (mQTL). Moreover, methylation levels at a specific chromosomal position may regulate gene expression levels (eQTM).
