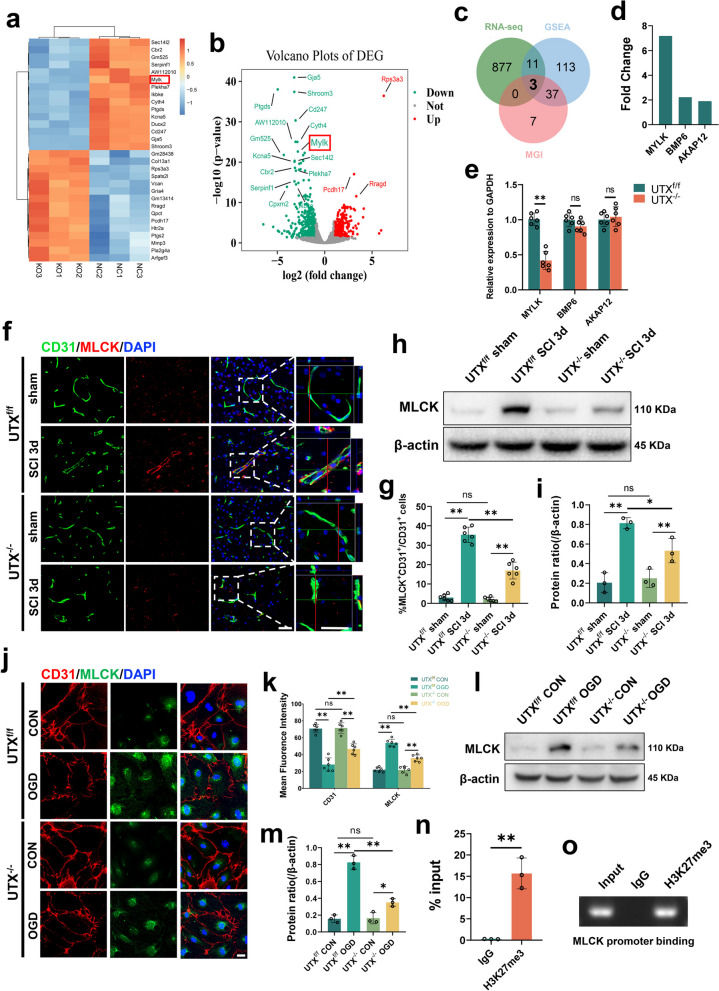
Fig. 5.
UTX plays a pivotal role in regulating MLCK expression. a, b UTXf/f SCMECs Vs UTX−/− SCMECs were significantly different (p ≤ 0.05, logFC ≥ 1 or ≤ − 1) from heatmap and volcano map of RNA expression profiles. c Venn diagram of the intersection of differential genes screened by RNA-seq and genes associated with vascular permeability retrieved from GSEA and MGI databases. d Differential ploidy of the genes screened in c. e qRT-PCR verification of the mRNA levels of MYLK (MLCK), BMP6, and AKAP12 in the UTXf/f and UTX−/− mice at 3 days post-SCI. n = 3 per group. f Representative immunofluorescence images of MLCK (red) and CD31 (green), DAPI (blue) staining of injured epicenter of spinal cord at SCI 3d. Scale bars, 50 μm. g Quantitative analysis of CD31+MLCK+ cells as a percentage of CD31+ cells in f, n = 6 per group. h Western blotting analysis of MLCK protein expression levels in UTXf/f and UTX−/− mice at sham and SCI 3d. i Quantitative analysis of the expression levels of MLCK in h. n = 3 per group. j Representative immunofluorescence images of MLCK (red) and CD31 (green), DAPI (blue) staining in UTXf/f and UTX−/− SCMECs after OGD. Scale bars, 20 μm. k Quantitative analysis of the fluorescence intensity of CD31 and MLCK in j, n = 6 per group. l Western Blotting analysis of MLCK protein expression levels in UTXf/f and UTX−/− SCMECs after OGD. m Quantitative analysis of the expression levels of MLCK in l. n = 3 per group. n ChIP-qPCR to detect the binding rate of histone H3K27me3 to MLCK initiation sequence. n = 3 per group. o Gel electrophoresis diagram of ChIP-qPCR products. Data are represented as mean ± SEM. ns P > 0.05, *P < 0.05, **P < 0.01