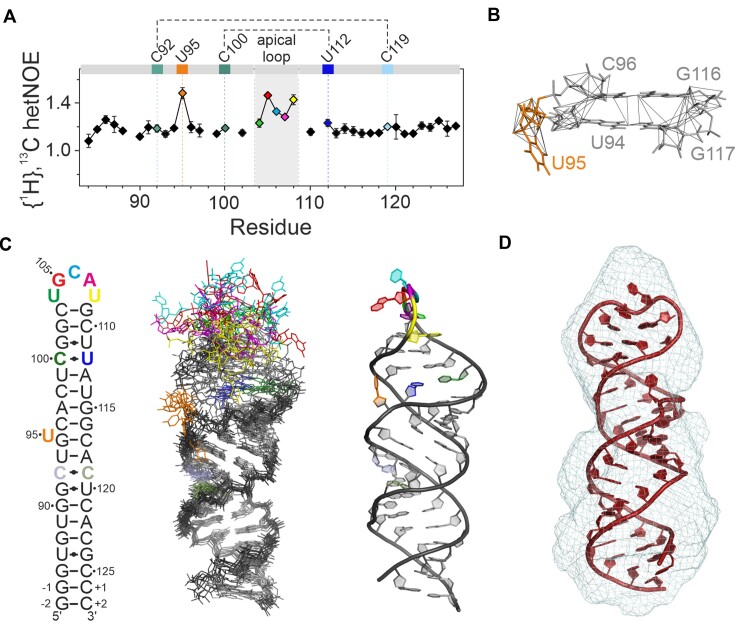
Figure 2.
NMR structure and structural details of 5_SL4. (A) {1H},13C-hetNOE of the aromatic pyrimidine H6C6 and purine H8C8 moieties of 5_SL4. Residues deviating from canonical A-form conformation are assigned and highlighted in different colors according to the color scheme in (C). Residues of the two pyrimidine•pyrimidine mismatches are connected by dotted lines. (B) NOE derived upper limit distance restraints of residue U95 (orange) and between its two adjacent base pairs (gray). The RNA is shown as a stick representation. The residues are assigned. Distance restraints are depicted as gray lines. (C) NMR structure of 5_SL4. Overlay of the 10 structures with the lowest target function (middle). The heavy atoms of all residues with H6/8C6/8 {1H},13C-hetNOE values <1.2 were used for the overlay. The RNA is shown as stick representation. Residues deviating from canonical A-form structure are highlighted in different colors according to the secondary structure (left). The structure with the lowest target function is shown to the right in cartoon representation displaying the same color scheme. (D) SAXS de novo envelope of 5_SL4 generated with the program DAMMIF superimposed with the best fitting NMR-structure (ninth lowest target function) using the program CIFSUB of the ATSAS v.3.1.3 software suite (38). The RNA is shown as a cartoon representation, the SAXS envelope as mesh.