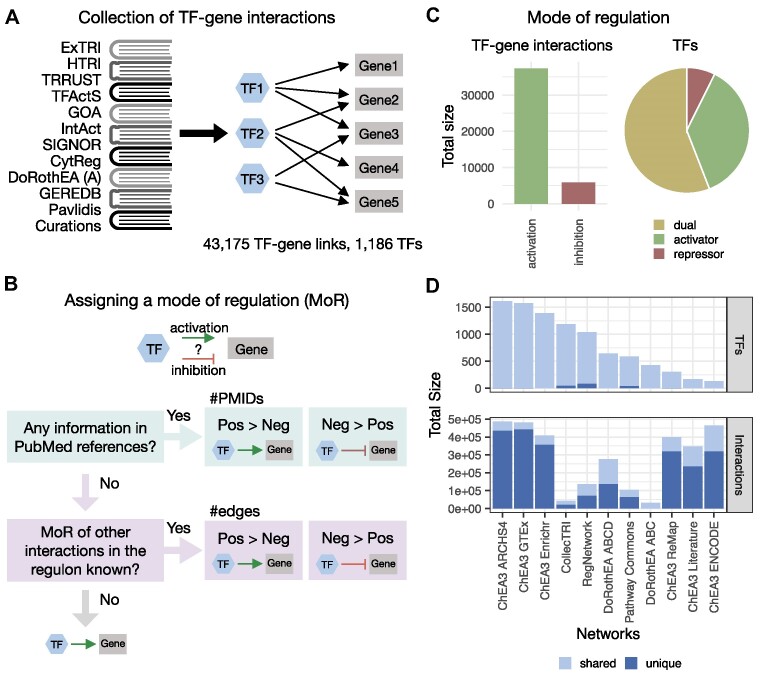
Figure 1.
Description of transcription factor (TF)–gene interactions in the CollecTRI-derived regulons and comparison to other regulon collections. (A) Collecting transcription factor (TF)–gene links to construct regulons from CollecTRI. Depicting prior knowledge resources used to collect links, which were aggregated within CollecTRI. (B) Flow chart describing how the mode of regulation (MoR) was assigned to each TF–gene link. The MoR, indicating the direction of transcriptional regulation from the TF to its target gene, was determined for each TF–gene link, based on factors such as PubMed references (PMIDs) and the MoR of other genes in the regulon. (C) Summary of the MoR for TF–gene interactions in CollecTRI. Total number of interactions for activating and repressive TF–gene links (left) and percentage of TFs that purely function as activators, repressors, or have a dual mode of regulation (right). (D) Comparison of the number of unique TFs (top) and interactions (bottom) across different resources—with ChEA3 ARCHS4, ChEA3 GTEx and ChEA3 Enrichr being solely based on co-expression or co-occurrence. Any TF or interaction present in more than one resource is considered shared.