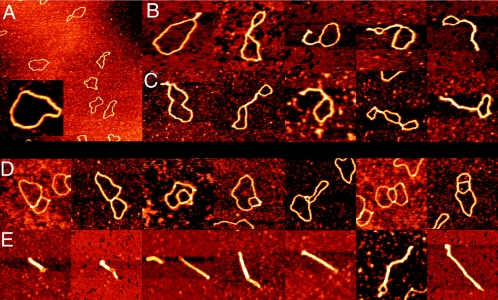
FIG. 2.
SFM images of circular nicked pUC19 DNA molecules with and without E. coli StpA or Pseudomonas strain Y1000 MvaT. (A) Bare relaxed pUC19 DNA molecules. Image shows a 3- by 3-μm surface area. (Inset) Close-up of a bare relaxed pUC19 DNA molecule at the same magnification as used in panels B to E. (B) DNA molecules after incubation with StpA (one dimer per 200 bp). Complexes are laterally compacted by antiparallel DNA bridging. (C) DNA molecules after incubation with MvaT (one dimer per 160 bp). Complexes are laterally compacted by antiparallel DNA bridging. (D) DNA molecules after incubation with MvaT (one dimer per 160 bp). Complexes are compacted by parallel DNA bridging. (E) DNA molecules after incubation with MvaT (one dimer per 80 bp). Two types of complexes (reflected in clearly different sizes) are formed. Large complexes are laterally fully compacted by regular antiparallel DNA bridging, and small complexes are more strongly compacted through extensive parallel loop-loop bridging. Images in panels B to E show an 0.5- by 0.5-μm surface area. Color represents height ranging from 0 to 2 nm.