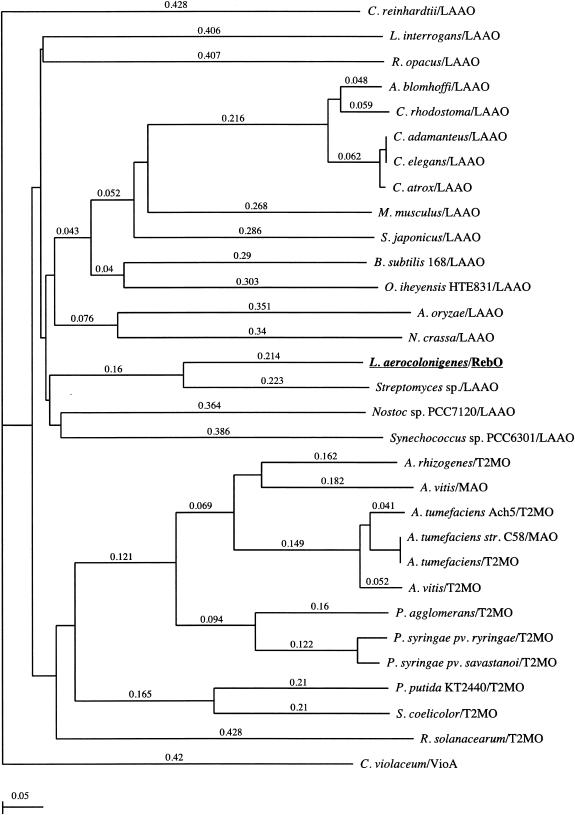
FIG. 3.
Phylogenetic tree representation of the apparent evolutionary distances of l-AAO (LAAO) proteins from several bacteria, fungi, and venomous snakes. Agkistroden blomhoffi LAAO, AB072392; Aspergillus oryzae LAAO, AB078782; Bacillus subtilis 168 LAAO, Z99114; Crotalus adamanteus LAAO, JE0266; Crotalus atrox LAAO, AF093248; Caenorhabditis elegans LAAO, NM_077599; Chlamydomonas reinhardtii LAAO, T08202; Calloselasma rhodostoma LAAO, P81382; Chromobacterium violaceum VioA, AF172851 and AB032799; Leptospira interrogans LAAO,AE011573; Mus musculus LAAO, BC017599 and NP_598653; Neurospora crassa LAAO, ABBX01000266; Nostoc sp. strain PCC7120 LAAO, NC_003276; Oceanobacillus iheyensis HTE831 LAAO, AP004597; Rhodococcus opacus LAAO, AY053450; S. japonicus LAAO, AJ400871; Streptomyces sp. LAAO, AB088119; Synechococcus sp. strain PCC6301 LAAO, Z48565; Agrobacterium tumefaciens strain C58 MAO, NC_003065; Agrobacterium vitis MAO, AF126447; Agrobacterium rhizogenes T2MO, Q09109; A. tumefaciens Ach5 T2MO, P04029; A. tumefaciens T2MO, NC_003308; A. vitis T2MO, P25017; Pantoea agglomerans T2MO, Q47861; Pseudomonas putida KT2440 T2MO, NC_002947; Pseudomonas syringae pv. syringae T2MO, A53376; P. syringae pv. savastanoi T2MO, A25493; Ralstonia solanacearum T2MO, NC_003295; S. coelicolor T2MO, AL939109. The tree was constructed by the neighbor-joining method using ClustalW (version 1.4). The scale for the tree indicates inferred evolutionary distances. MAO, monoamine oxidase; T2MO, tryptophan 2-monooxygenase.