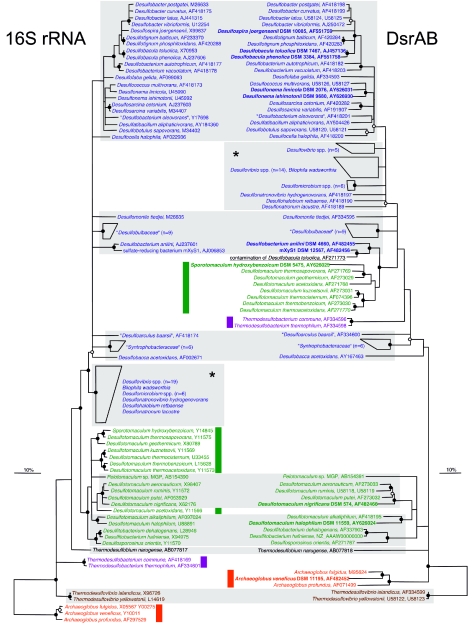
FIG. 2.
Comparison of 16S rRNA- and DsrAB-based phylogenetic consensus trees. Sequences determined in this study are in boldface. 16S rRNA phylogenetic analyses were performed on alignment positions conserved in at least 50% of all bacteria. Alignment regions of insertions and deletions were omitted in DsrAB amino acid sequence analyses. Polytomic nodes connect branches for which a relative order could not be determined unambiguously by using distance matrix, maximum-parsimony, and maximum-likelihood methods. Filled circles indicate branch points highly supported by maximum-parsimony bootstrap analysis (>90% in 1,000 resamplings). Open circles at nodes indicate 75 to 90%, while nodes without circles showed <75% bootstrap support. The bars represent 10% sequence divergence as estimated from maximum-likelihood and distance matrix analysis for the 16S rRNA and DsrAB trees, respectively. “Deltaproteobacteria,” low G+C gram-positive bacteria (Firmicutes), Thermodesulfobacterium species, Thermodesulfovibrio species, Thermodesulfobium narugense, and Archaeoglobus species are depicted in blue, green, violet, brown, black, and red, respectively. The wrong dsrAB sequence of D. toluolica published by Klein et al. (15) is underlined. Colored bars indicate species which harbor laterally acquired dsrAB genes. Consistent groups between both trees are shaded gray. Note that the apparently inconsistent positions of SRP groups that are labeled by an asterisk are not well resolved in the respective trees and thus cannot be interpreted as indicators of lateral gene transfer events. The strain A. veneficus SNP6 (DSM 11195) (containing plasmid XY), had been deposited in the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) by K. O. Stetter, Lehrstuhl für Mikrobiologie, Universität Regensburg, Regensburg, Germany. An ungrouped version of this figure (supplementary web Fig. 1) can be downloaded from our web site (http://www.microbial-ecology.net/supplements.asp) together with the respective ARB dsrAB database.