Abstract
Agrobacterium tumefaciens stands as one of biotechnology's greatest successes, with all plant genetic engineering building on the strategies of this pathogen. By integrating responses to external pHs, phenols, and monosaccharides, this organism mobilizes oncogenic elements to efficiently transform most dicotyledonous plants. We now show that the complex signaling network used to regulate lateral gene transfer can be resolved as individual signaling modules. While pH and sugar perception are coupled through a common pathway, requiring both low pH and sugar for maximal virulence gene expression, various VirA and ChvE alleles can decouple pH and monosaccharide perception. This VirA and ChvE system may represent a common mechanism that underpins external pH perception in prokaryotes, and the use of these simple genetic elements may now be extended to research on specific responses to changes in environmental pH.
The hydrogen ion concentration represents a critical environmental constraint for all macromolecular structures, physiological networks, and biological energy conversions. Accordingly, the ability to sense and respond to changes of pH is essential for all organisms. In contrast to the vast number of genes discovered to be pH regulated, the actual proteins responsible for sensing H+ are poorly defined. Only a few systems, such as the PhoQ/PhoP system in Salmonella enterica serovar Typhimurium (3), the ChvG/ChvI system in Agrobacterium sp. (26), and the chemotactic proteins Tar and Tsr (43), are known to be involved in sensing either extracellular or intracellular pH. Enteric bacteria, such as Escherichia coli and S. enterica serovar Typhimurium, have evolved sophisticated acid tolerance response systems to adapt to adverse pH environments (4, 18), but the pH sensing mechanisms remains ill defined. This deficiency both hinders development of a system-wide understanding of pH responses and limits our capability to further regulate and exploit these elements as pH sensors.
The response to extracellular pH is particularly critical for many pathogenic and symbiotic relationships in which a change in hydrogen ion concentration can mark a transition from a free-living mode to the host environment. Agrobacterium tumefaciens, a soil pathogen that causes plant tumors via the transfer of oncogenic DNA into host cells, uses pH as a part of its arsenal of host indicators. Low pH (approximately 4.8 to 5.5), monosaccharides (including glucose), and phenols (e.g., acetosyringone [AS]) are all required to maximally induce the expression of the virulence (vir) genes that mediate the transfer of DNA into the plant genome (6, 42, 54). Two genes, the VirA/VirG two-component sensor-transducer system, regulate the production of both the transferred oncogenic DNA and the DNA transfer machinery from the tumor-inducing Ti plasmid (22, 50, 52). VirA, a membrane-localized histidine sensor kinase, is autophosphorylated upon perceiving signals characteristic of host wound sites and transfers the phosphoryl group to VirG (32). Phosphorylated VirG acts as a transcription activator that induces the expression of the remaining genes of the vir regulon.
VirA contains two membrane-spanning segments, a periplasmic (P) domain, and a large cytoplasmic C terminus that has been subdivided into linker (L), kinase (K), and receiver (R) domains (12). The linker domain has been implicated in phenol sensing (12). Monosaccharides are perceived through the periplasmic domain of VirA via chromosomal sugar binding protein ChvE, and their perception enhances the sensitivity and maximal response to phenolic compounds (1, 37, 38). Optimal vir gene induction, however, does not occur at neutral pHs (40) but rather requires an acidic environment. Part of this external pH dependence is attributed to enhanced virG expression, as mediated by the acidic pH-inducible P2 promoter (13, 28, 49). However, replacing wild-type P1 and P2 promoters of virG with the lac promoter does not create a pH-independent virulence response (15), indicating that other pH-regulated inputs are mediated through the VirA/VirG system.
Chang and Winans (12) demonstrated that the kinase domain of VirA could activate vir gene expression independent of pH, while VirA truncation mutants with only the linker and kinase domains were still pH responsive. They concluded that the cytoplasmic linker domain of VirA was responsible for pH input. However, a later report (14) showed that an allele product of VirA, of which the linker domain was truncated, maintaining the periplasmic and kinase domains, still responded to a low-pH stimulus. Other investigators constructed fusion proteins with the periplasmic domain replaced by one of the E. coli membrane chemoreceptors, Tar (30). One of the hybrid proteins achieved high induction even at pH 7.0, further suggesting that the periplasmic domain of VirA is critical for pH sensing. Here we exploit the modularity of the histidine sensory kinases to establish that the periplasmic domain of VirA is involved in pH perception. Further, we show that the pH response is directly coupled with sugar signaling through ChvE-VirA association. This knowledge allows the multiple inputs to be separated and several alleles of ChvE and VirA to be developed as pH-specific response modules.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. E. coli strains XL1-Blue and DH5α were used for routine cloning. A. tumefaciens strains were grown at 28°C in Luria-Bertani (LB) medium or induction medium (11) containing either glucose or glycerol. The chvE deletion strain, DL8, was constructed as follows. The chvE gene was cloned from genomic DNA of A136 by PCR amplification and then inserted into pUC19 to create pRG157. A kanamycin resistance gene was then cloned from pUC4K (Pharmacia) into the BamHI site within the chvE gene of pRG157. The resulting plasmid, pRG159, was electroporated into A136. The correct marker exchange mutant, DL8, was selected from kanamycin-resistant and ampicillin-sensitive colonies and further confirmed by colony PCR.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacIqZΔM15 Tn10 (Tcr)] | Stratagene |
| DH5α | recA1 endA1 hsdR17 supE44 gyrA96 relA1Δ(lacZYA-argF)U169 (φ80dlacZΔM15) | Invitrogen |
| A. tumefaciens | ||
| A348 | A136 containing pTiA6NC | 19 |
| A136 | Strain C58 cured of pTi plasmid | 48 |
| DL8 | A136 chvE deletion, Kmr | This study |
| Plasmids | ||
| pBluescript II KS(+/−) | Cloning vector, ColE1 Apr | Stratagene |
| pJZ4 | 5.1-kb fragment containing PvirB-lacZ-nptII cloned into pMON596, Inc P Specr | 53 |
| pRG109 | PN25-His6-virG in pJZ4, Specr | This study |
| pRG108 | PN25-His6-virG(N54D) in pJZ4, Specr | This study |
| pYW47 | PN25-His6-virG in pYW15, IncW Apr | 47 |
| pAM13 | PN25-His6-virG(N54D) in pYW15, IncW Apr | 32 |
| pSM102 | occQ::lacZ, IncP Apr | 39 |
| pVRA8 | virA from pTiA6 in pUCD2, pBR322ori, IncW Apr | 25 |
| pMutA | virA(G665D) in pUCD2, pBR322ori, IncW Apr | 29 |
| pYW15 | Broad-host-range expression vector, IncW Apr | 47 |
| pFF4 | PN25-virA in pQE50, ColE1 Apr | This study |
| pFQ25 | PN25-virA and PN25-virG, IncW Apr | This study |
| pYW21 | PN25-His6-virA(285-829) in pYW15, Apr | 45 |
| pYW39 | PN25-His6-virA(285-829)(G665D) in pYW15, Apr | 47 |
| pAM23 | PN25-His6-virA(285-711) in pYW15, Apr | 32 |
| pAM28 | PN25-His6-virA(285-711) (G665D) in pYW15, Apr | 32 |
| pRG100 | PN25-His6-virA(426-829) (G665D) in pYW15, Apr | This study |
| pRG118 | PN25-His6-virA(426-711) (G665D) in pYW15, Apr | This study |
| pRG119 | PN25-His6-virA(426-711) in pYW15, Apr | This study |
| pRG120 | PN25-His6-virA(426-829) in pYW15, Apr | This study |
| pRG135 | virA(1-711) in pYW15, Apr | This study |
| pRG157 | chvE from A136 in pUC19, ColE1 Apr | This study |
| pRG159 | Kmr gene cloned in BamH I site of chvE in pRG157, ColE1 Apr Kmr | This study |
| pRG162 | virA(E210V) in pBluescript II KS (+/−), ColE1 Apr | This study |
| pRG165 | PN25-chvE in pYW15, Apr | This study |
| pRG166 | PN25-chvE(T211M) in pYW15, Apr | This study |
| pRG168 | virA and PN25-chvE in pYW15, Apr | This study |
| pRG169 | virA and PN25-chvE(T211M) in pYW15, Apr | This study |
| pRG170 | virA(E210V) and PN25-chvE in pYW15, Apr | This study |
| pRG171 | virA(E210V) and PN25-chvE(T211M) in pYW15, Apr | This study |
Plasmid constructions.
The plasmids used in this study are listed in Table 1. The coliphage T5 promoter (PN25) was used to drive the expression of virG independent of environmental factors like pH and phosphate concentration (47). The NcoI fragments containing either PN25-His6-virG (where His6 stands for a six-histidine tag) from pYW47 or PN25-His6-virG(N54D) from pAM13 were treated with Klenow fragment and ligated with pJZ4 (PvirB-lacZ) that was treated with KpnI and Klenow fragment, resulting in pRG109 and pRG108.
The coding region of the virA gene was placed behind PN25 to create pFF4, and PN25-virA was subsequently released for insertion into a pYW47 derivative plasmid to give pFQ25. The VirA mutant construct comprising amino acids (aa) 426 through 829 with a G665D mutation in the kinase domain [VirA(426-829)(G665D)] was amplified by PCR from pYW39 with primers 5′-ATTCAGCTTCTTGAACTCGCCACC-3′ and 5′-GCGGTACCCTACGTCTTGATTTTGGTTAG-3′ (KpnI), followed by digestion with KpnI and ligation with EcoICRI- and KpnI-digested pYW15 to generate pRG100. pRG118, pRG119, and pRG120 were made by replacing the HindIII fragment of pRG100 with HindIII fragments from pAM23 (coding for stop codon after aa 711), pAM28 (coding for wild-type kinase domain and stop codon after aa 711), and pYW45 (coding for wild-type kinase and receiver domains of VirA) (46), respectively. The VirA with the truncated receiver domain (aa 1 to 711) was released from pCH355 (14) as a KpnI fragment and inserted into KpnI-digested pYW15 to make pRG135.
To place chvE behind the PN25 promoter, chvE was amplified from pRG157 by PCR oligonucleotides 5′-CACAGAATTCATTAAAGAGGAGAAATTAACTATGAAGTCCATTATTTCG-3′ (EcoRI) and 5′-CGTCTTGGTGATGTTCCGCATTTC-3′ and cloned into pCR2.1-TOPO by TOPO cloning (Invitrogen). Subsequently, chvE was released as an EcoRI fragment and ligated into pYW15 to create pRG165. For site-directed mutagenesis of virA(E210V) and chvE(T211M), the QuickChange site-directed mutagenesis kit (Stratagene) was used. Primers 5′-CGCATACTTGCACGTGTAGGTCCCATTATCTTATCG-3′ and 5′-CGATAAGATAATGGGACCTACACGTGCAAGTATGCG-3′ were used to introduce the E210V mutation into the virA gene, resulting in pRG162. Primers 5′-GAGCCTGGGCCATTGCCGGATCCC-3′ and 5′-GGGATCCGGCAATGGCCCAGGCTC-3′ were used to introduce the T211M mutation into chvE of pRG165, creating pRG166. The sequences of all PCR products were confirmed by DNA sequencing. The wild-type virA gene and virA(E210V) were cut from pVRA8 and pRG162 by use of KpnI and cloned into the KpnI sites of pRG165 and pRG166 to generate pRG168, pRG169, pRG170, and pRG171.
Immunoblot analysis.
A. tumefaciens cells were grown overnight in 30 ml of LB medium with the appropriate antibiotics. The bacterial pellets were washed with phosphate-buffered saline and lysed by brief sonication on ice. Clarified lysates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in polyacrylamide gels (Invitrogen), followed by electro-blotting onto nitrocellulose membranes using the Mini Trans-Blot transfer apparatus (Bio-Rad). Visualization of His6-tagged proteins was achieved by Western blot using anti-RGS-His monoclonal antibody (QIAGEN).
β-Galactosidase assays for vir gene induction.
pRG109 or pRG108 carrying the β-galactosidase reporter PvirB-lacZ and PN25-virG or PN25-virG(N54D) was used to assay vir gene expression in Ti-cured Agrobacterium strains. Cells were grown in 20 ml of LB medium to an optical density at 600 nm (OD600) of 0.4 to 0.6 in the presence of the appropriate antibiotics. The cell mass was pelleted by centrifugation for 10 min at 7,000 × g at 4°C and washed with phosphate-buffered saline. The pellet was diluted to an OD600 of ∼0.1 into tubes containing a total of 1 ml of induction media (50 mM MES [2-(4-morpholino)-ethanesulfonic acid], 1× AB salts) supplemented with 1% glycerol (11) and cultured at 28°C with shaking at 225 rpm for 15 h. β-Galactosidase activity was determined as described previously (31) with readings of optical densities at 600 and 415 nm using the EL800 microplate reader (BIO-TEK Instruments).
RESULTS
virA truncation mutants.
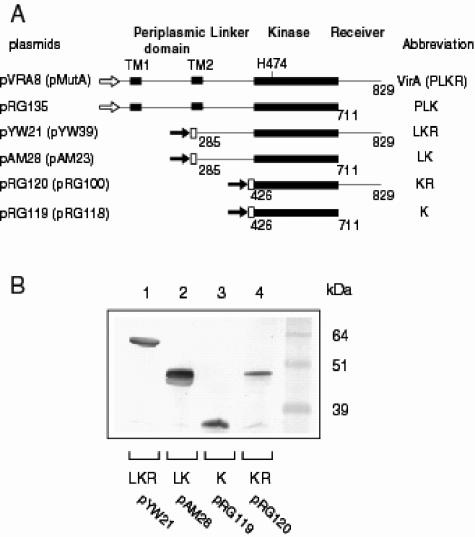
virG was placed behind a constitutive PN25 promoter (47) to eliminate pH activation of the P2 virG promoter (28, 49). pRG109, containing PN25-virG and PvirB-lacZ, together with plasmids expressing various virA truncation mutants, was transformed into A. tumefaciens A136, a strain lacking pTi, to assay for lacZ expression. Among the constructs evaluated in Fig. 1A, VirA(285-829) (LKR), VirA(285-711) (LK), VirA(426-829) (KR), and VirA(426-711) (K) were fused with an N-terminal His6 tag. Western blot analyses using anti-His monoclonal antibody showed that all these VirA truncation proteins were stably expressed (Fig. 1B) and the change of pH from 5.5 to 7.0 did not alter the levels or apparent stability of these proteins (data not shown).
FIG. 1.
Protein expression of VirA truncation mutants. (A) Schematic representation of VirA constructs. The plasmids expressing VirA constructs are listed on the left; in parentheses are the corresponding plasmids of VirA constructs with a G665D mutation in the kinase domain. Open arrow, virA promoter; solid arrow, PN25 promoter; white box, His6 tag. (B) Western blot analysis of VirA constructs. Clarified lysates from bacteria carrying indicated plasmids were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in 10% Bis-Tris NuPage (Invitrogen) gels followed by Western blot analysis using anti-RGS-His monoclonal antibody.
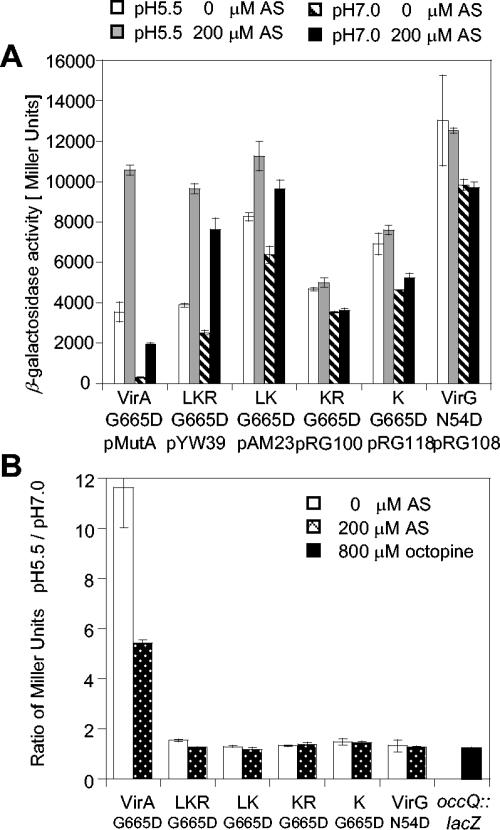
The pH- and AS-dependent responses of virA allele products are shown in Fig. 2A. Consistent with previous reports implicating the linker domain's involvement in AS sensing (12, 14), the kinase alone (K) displayed AS-independent activity while LKR and LK remained AS-inducible. When the pH was reduced from 7.0 to 5.5, all strains showed an increase of vir expression, but a far greater increase was observed for the full-length VirA strain. Figure 2B presents pH responses as ratios of the activities at pH 5.5 and pH 7.0. The pH 5.5/pH 7.0 ratio of the full-length VirA strain was around 10, while VirA mutants lacking the periplasmic domain (LKR, LK, KR, and K) had ratios between 1.2 and 2.0. The pH ratio appeared to be independent of the promoter, as both autoinducing wild type and PN25 had similar increases. Moreover, removal of the inhibitory receiver domain (12, 14) (PLK) significantly elevated both the basal and induced activity (Fig. 2A) beyond the linear range of the β-galactosidase assay under these conditions. Here the pH 5.5/pH 7.0 ratio was at least 6, significantly higher than the ratios for LKR, LK, KR, and K, strongly supporting a role of the periplasmic domain in pH sensing.
FIG. 2.
pH responses of VirA constructs. A. tumefaciens A136 carrying the indicated VirA constructs together with pRG109 containing PvirB-lacZ and PN25-virG was cultured in induction medium supplemented with 14 mM glucose and assayed for β-galactosidase activity. (A) Expression of PvirB-lacZ at pH 5.5 with 0 μM AS, pH 5.5 with 200 μM AS, pH 7.0 with 0 μM AS, and pH 7.0 with 200 μM AS. (B) Ratio of the β-galactosidase activity, pH 5.5/pH 7.0.
Therefore, all the periplasmic-domain-truncated variants of VirA displayed a much smaller pH 5.5/pH 7.0 ratio around 2. This weak pH response may be consistent with the previous conclusion that linker-domain-containing constructs (LKR and LK) still responded to low pH (12). However, the octopine reporter occQ::lacZ (39) and the product of the VirG(N54D) allele, which activates vir expression independent of either VirA or AS (21, 35), also showed a similar twofold increase (Fig. 3). Thus, this weak pH response appears not specific for VirA but more consistent with a small global pH effect, possibly reflecting the overall control of expression from pTi during pathogenesis. Further, the products of VirA alleles carrying a G665D substitution, which are known to give both basal and AS-inducible vir expressions (29), had pH responses similar to those of the wild-type VirA variants, for which a high pH 5.5/pH 7.0 ratio was associated only with VirA allele products containing an intact periplasmic domain (Fig. 3).
FIG. 3.
pH response of VirA(G665D) constructs. A. tumefaciens A136 carrying the indicated VirA constructs and pRG109 was cultured as described above in the presence of 14 mM glucose. (A) Expression of PvirB-lacZ at pH 5.5 with 0 μM AS, pH 5.5 with 200 μM AS, pH 7.0 with 0 μM AS, and pH 7.0 with 200 μM AS. (B) Ratio of the β-galactosidase activity at pH 5.5 to that at pH 7.0 with 0 and 200 μM AS.
Interdependence of pH and sugar perception.
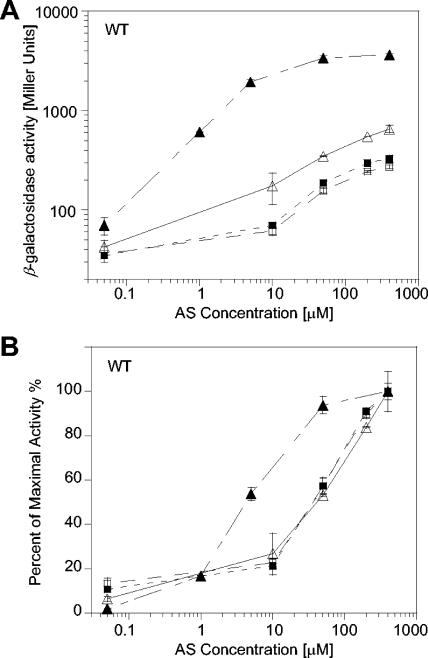
Analysis of these VirA truncation allele products implicated a common role for the periplasmic domain in pH and monosaccharide perception. To investigate whether these were distinct or interdependent perception events, cells containing wild-type VirA were induced with the indicated concentrations of AS at pH 5.5 and 7.0 in the presence and absence of 13 mM glucose (Fig. 4). Glucose enhanced the maximal induction at pH 5.5 >5-fold; however, this increase was dependent on both glucose and acidic pH (Fig. 4A). At pH 7.0, no sugar-enhanced increase was observed; without glucose, the maximal induction by an acidic pH was twofold, consistent with the global pH dependence observed in Fig. 2. In addition, glucose perception has been associated with an increase in both the maximal induction and the sensitivity of the VirA/VirG system to AS (2). In Fig. 4B, the dosage-dependent data plotted in Fig. 4A are expressed as percentages of maximal activity in order to emphasize this sensitivity difference. A 10-fold shift of AS sensitivity was apparent only when both glucose and acidic pH were present.
FIG. 4.
PvirB-lacZ expression by wild-type VirA as a function of AS concentrations. A. tumefaciens A136/pRG109 containing pVRA8 was cultured for 16 h in media supplemented with 1% glycerol at pH 5.5 (Δ), pH 5.5 with 14 mM glucose (▴), pH 7.0 (□), and pH 7.0 with 14 mM glucose (▪). β-Galactosidase activity calculated as Miller units (A) and expressed as percentages of the maximal activity (B).
The periplasmically localized sugar binding protein ChvE is required for glucose sensing in Agrobacterium (10, 37). Disruption of chvE in mutant strain DL8 abolished the pH or sugar response (Fig. 5A and B). When PN25-chvE was moved into strain DL8, the maximal induction (Fig. 5C) and AS sensitivity (Fig. 5D) were both rescued, but this was the case only when both glucose and acidic-pH stimuli were present. Given the similar responses to pH and sugar for A136 carrying a wild-type chvE (Fig. 4) and DL8/pRG168 carrying PN25-chvE (Fig. 5C and D), the wild-type chvE promoter was not critical for coupling the responses to pH and sugar. Taken together, these data suggest that pH and sugar sensing are mechanistically coupled.
FIG. 5.
PvirB-lacZ expression by wild-type VirA as a function of AS concentrations in a background in which chvE was disrupted. A. tumefaciens DL8/pRG109 containing pVRA8 (A and B) or pRG168 (C and D) was cultured as described at pH 5.5 (Δ), pH 5.5 with 14 mM glucose (▴), pH 7.0 (□), and pH 7.0 with 14 mM glucose (▪). β-Galactosidase activity calculated as Miller units (A and C) and expressed as percentages of the maximal activity (B and D).
Decoupling pH and sugar perception.
Genetic evidence provides the strongest support for a direct interaction between ChvE and the periplasmic domain of VirA. VirA allele products containing an E210V substitution in the periplasmic domain are not responsive to glucose (37, 41), but a compensating ChvE(T211M) allele product was discovered that fully restored sugar perception (37). Consistent with these reports, VirA(E210V) didn't respond to glucose, even with PN25-chvE, while DL8/pRG171 containing VirA(E210V) and PN25-chvE(T211M) had a wild-type pH or sugar response (data not shown), mediating both an increased maximal induction and an enhanced AS sensitivity.
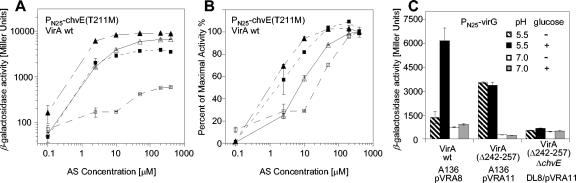
While the precise arrangement of these compensating interactions between VirA and ChvE remains unclear in the absence of greater structural resolution, the importance of these residues to perception motivated investigation of the pH or sugar response of wild-type VirA with PN25-chvE(T211M). DL8/pRG169, expressing the wild-type VirA and ChvE(T211M), displayed maximal induction at pH 5.5 without glucose (Fig. 6A). The acidic pH alone induced a >10-fold increase over pH 7.0 induction. Glucose alone was also sufficient to increase the maximal activity at a neutral pH; similar increases in AS sensitivity are also apparent in Fig. 6B. Without glucose at pH 7.0, the AS sensitivity of VirA with ChvE(T211M) was similar to that of VirA with the wild-type ChvE as shown in Fig. 5D. Therefore, either adding glucose or dropping the pH to 5.5 increased AS sensitivity. The pH and sugar responses are no longer coupled for these allele products; either glucose or low pH alone is a sufficient inducer signal.
FIG. 6.
pH and sugar responses of VirA and ChvE allele products. PvirB-lacZ expression by DL8/pRG109 containing pRG169 with wild-type VirA and PN25-chvE(T211M) at pH 5.5 (Δ), pH 5.5 with 14 mM glucose (▴), pH 7.0 ([squo]), and pH 7.0 with 14 mM glucose (▪), in terms of β-galactosidase activity calculated as Miller units (A) and expressed as percentages of the maximal activity (B). (C) PvirB-lacZ expression by wild-type VirA and VirA with aa 242 through 257 deleted (Δ242-257) with 200 μM AS.
Residues of VirA outside of the predicted ChvE binding site have also been implicated in sugar signaling. For example, allele products carrying E255Q or lacking amino acids 242 through 257 of VirA are no longer sugar responsive (2). In strain A136 carrying a wild-type copy of chvE, VirA with aa 242 through 257 deleted was not responsive to glucose, but the activity at pH 5.5 was more than 10-fold higher than that at pH 7.0 (Fig. 6C). When this VirA allele is placed in the DL8 strain, in which chvE is disrupted, pH induction is no longer observed. Therefore, the pH response is dependent not on sugar but on chvE achieving a highly specific, sugar-independent pH response element.
DISCUSSION
Environmental pH sensing represents a critical regulatory module that interconnects with virtually every component of the cellular network, controlling responses as diverse as cell growth, energy interconversion, taxis, symbiosis, and pathogenesis. Pathogens have evolved simple, efficient and, very precise host sensing modules (33). For example, the tumor-inducing plasmid of Agrobacterium encodes a two-component sensor kinase, VirA, which mediates virulence by responding to environmental pH. While VirA responds to diverse structural inputs, we have exploited the modularity of this two-component sensor kinase to resolve the pH-signaling module and generate elements that respond specifically to extracellular pH.
Agrobacterium is shown here to maintain a global acidic pH response, generally increasing pTi gene expression by approximately 1.2- to 2.0-fold when the environmental pH drops from 7.0 to 5.5. Acidic pH is known to increase the expression of virG through the P2 promoter (49, 51), and the previously discovered ChvG/ChvI two-component system (26) may well play an important role in promoter activation (13), but little is currently known about its mechanism. This global pH response may also have complicated previous assignments of the cytoplasmic domain of VirA in pH sensing (12).
In addition to this global pH response, distinct pH activation is associated specifically with the periplasmic domain of VirA. Removal of the periplasmic domain elevated vir induction over that of full-length VirA at pH 7.0, consistent with the periplasmic domain limiting the activity of VirA at pH 7.0. Banta et al. and others showed that the removal of aa 63 through 240, the presumed sugar binding region in the periplasmic domain, resulted in higher vir expression in the absence of sugars, supporting a model in which the ChvE-sugar complex relieves repression by the periplasmic domain (2, 10, 38). Thus, the periplasmic domain of VirA may play an inhibitory role, and the perception of sugars and pH relieves this inhibition to achieve the host-specific response.
This pH response is mechanistically coupled with sugar perception. Either the absence of sugars (Fig. 4) or the disruption of chvE (Fig. 5A and B) abolishes pH signaling, a response rescued by the in trans expression of chvE (Fig. 5C and D). Such pH and sugar perceptions may be coupled by either pH-induced expression of chvE or a conformation change required for optimal ChvE-VirA interaction. chvE expression is regulated by a subset of inducing sugars through GbpR, a LysR family transcriptional regulator (16, 36), and there is no evidence for the existence of a pH-regulated promoter for either chvE or gbpR. Moreover, replacing the wild-type chvE promoter with a constitutive PN25 promoter does not decouple pH sensing and sugar sensing. However, we cannot exclude the possibility that extracellular pH alters the turnover of the periplasmic ChvE protein, resulting in the dependence of sugar response on environmental pH.
Specific substitutions in ChvE override the requirement for both low pH and sugar for optimal vir expression. The ability of ChvE(T211M) to rescue the sugar response of VirA(E210V) might suggest that residue 211 is critical to the VirA-ChvE interaction, possibly by increasing the binding affinity to VirA so that either sugar- or low-pH-induced changes are sufficient for perception. Therefore, pH or sugar sensing is most likely mediated through a direct interaction between VirA and ChvE.
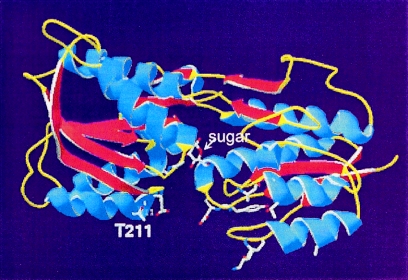
Figure 7 shows the homology model built on the crystal structure templates of ribose binding protein (RBP) and glucose-galactose binding protein (GBP). T211 is predicted to reside on the surface of ChvE, close to the “lips” of two domains in RBP and GBP where mutations that alter the protein-protein interactions with chemoreceptors and membrane transport proteins have been found (5, 7, 17). The binding affinity of sugar substrates can also be altered by mutations in this region, even though it is distinct from the buried sugar binding site (34, 44). Moreover, GalR, one of the LacI-GalR family of bacterial repressor proteins that has an N-terminal DNA binding domain and a C-terminal domain homologous to those of sugar binding proteins, displayed both pH- and galactose-dependent behavior of DNA binding (8). Further analysis suggested that pH and galactose could both modify the structure of the sugar binding domain of GalR to affect dimer formation and alter DNA binding affinity (9, 23). In addition, CcpA, another member of the GalR family of proteins, requires both an acidic pH and its sugar substrate glucose-6-phosphate to bind its regulatory DNA element (20). Therefore, it is not surprising that both a certain pH and sugars are required to activate ChvE for optimal interaction with VirA.
FIG. 7.
Simulated structure of ChvE. The structural data from GBP and RBP were used as templates to generate the coordinates for ChvE by use of Swiss-Prot. The resulting structure of ChvE contains two domains joined by three hinges with the sugar binding site located within the domain cleft. Based on the RBP and GBP models, the indicated side chains are involved in protein-protein interactions.
Taken together, pH sensing through VirA relies on the VirA-ChvE interaction and is coupled with the perception of other signals. This pH sensing mechanism, the first to be described in microorganisms, may be common. For example, both external pH and iron are sensed by the PmrA/PmrB two-component system of plant pathogen Erwinia carotovora subsp. carotovora to contribute to bacterial virulence (24). This ability to switch on only in the presence of two signals could greatly increase response precision. Similar arguments have been made for phenol perception by VirA (46), in which multiple inputs “ratchet” in the full response. However, the additive response of the VirA-ChvE interaction may not be solely at the level of perception. Removal of aa 242 through 257 from VirA leads exclusively to a pH sensor. This deletion could alter the ChvE binding site, decoupling pH and sugar perception, but the amino acids deleted abut the second membrane-spanning segment of VirA, suggesting that signal transmission through the membrane may be altered. Thus, sugar and pH transmission events may be processed separately. We are now positioned to resolve these mechanistic questions, to extend the use of these signaling modules in heterologous hosts (27), and to evolve different pH responses and alternate signal inputs for other regulatory networks.
Acknowledgments
We thank Andrew Binns and Arlene Wise in the Department of Biology, University of Pennsylvania, for discussions and suggestions on the manuscript.
This study was supported by National Institutes of Health grant GM47369.
REFERENCES
- 1.Ankenbauer, R. G., and E. W. Nester. 1990. Sugar-mediated induction of Agrobacterium tumefaciens virulence genes: structural specificity and activities of monosaccharides. J. Bacteriol. 172:6442-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banta, L. M., R. D. Joerger, V. R. Howitz, A. M. Campbell, and A. N. Binns. 1994. Glu-255 outside the predicted ChvE binding site in VirA is crucial for sugar enhancement of acetosyringone perception by Agrobacterium tumefaciens. J. Bacteriol. 176:3242-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bearson, B. L., L. Wilson, and J. W. Foster. 1998. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J. Bacteriol. 180:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bearson, S., B. Bearson, and J. W. Foster. 1997. Acid stress responses in enterobacteria. FEMS Microbiol. Lett. 147:173-180. [DOI] [PubMed] [Google Scholar]
- 5.Binnie, R. A., H. Zhang, S. Mowbray, and M. A. Hermodson. 1992. Functional mapping of the surface of Escherichia coli ribose-binding protein: mutations that affect chemotaxis and transport. Protein Sci. 1:1642-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binns, A. N. 2002. T-DNA of Agrobacterium tumefaciens: 25 years and counting. Trends Plant Sci. 7:231-233. [DOI] [PubMed] [Google Scholar]
- 7.Bjorkman, A. J., R. A. Binnie, H. Zhang, L. B. Cole, M. A. Hermodson, and S. L. Mowbray. 1994. Probing protein-protein interactions. The ribose-binding protein in bacterial transport and chemotaxis. J. Biol. Chem. 269:30206-30211. [PubMed] [Google Scholar]
- 8.Brenowitz, M., E. Jamison, A. Majumdar, and S. Adhya. 1990. Interaction of the Escherichia coli Gal repressor protein with its DNA operators in vitro. Biochemistry 29:3374-3383. [DOI] [PubMed] [Google Scholar]
- 9.Brown, M. P., N. Shaikh, M. Brenowitz, and L. Brand. 1994. The allosteric interaction between d-galactose and the Escherichia coli galactose repressor protein. J. Biol. Chem. 269:12600-12605. [PubMed] [Google Scholar]
- 10.Cangelosi, G. A., R. G. Ankenbauer, and E. W. Nester. 1990. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc. Natl. Acad. Sci. USA 87:6708-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cangelosi, G. A., E. A. Best, G. Martinetti, and E. W. Nester. 1991. Genetic analysis of Agrobacterium. Methods Enzymol. 204:384-397. [DOI] [PubMed] [Google Scholar]
- 12.Chang, C. H., and S. C. Winans. 1992. Functional roles assigned to the periplasmic, linker, and receiver domains of the Agrobacterium tumefaciens VirA protein. J. Bacteriol. 174:7033-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, C. H., and S. C. Winans. 1996. Resection and mutagenesis of the acid pH-inducible P2 promoter of the Agrobacterium tumefaciens virG gene. J. Bacteriol. 178:4717-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, C. H., J. Zhu, and S. C. Winans. 1996. Pleiotropic phenotypes caused by genetic ablation of the receiver module of the Agrobacterium tumefaciens VirA protein. J. Bacteriol. 178:4710-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, C. Y., and S. C. Winans. 1991. Controlled expression of the transcriptional activator gene virG in Agrobacterium tumefaciens by using the Escherichia coli lac promoter. J. Bacteriol. 173:1139-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doty, S. L., M. Chang, and E. W. Nester. 1993. The chromosomal virulence gene, chvE, of Agrobacterium tumefaciens is regulated by a LysR family member. J. Bacteriol. 175:7880-7886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eym, Y., Y. Park, and C. Park. 1996. Genetically probing the regions of ribose-binding protein involved in permease interaction. Mol. Microbiol. 21:695-702. [DOI] [PubMed] [Google Scholar]
- 18.Foster, J. W. 1999. When protons attack: microbial strategies of acid adaptation. Curr. Opin. Microbiol. 2:170-174. [DOI] [PubMed] [Google Scholar]
- 19.Garfinkel, D. J., R. B. Simpson, L. W. Ream, F. F. White, M. P. Gordon, and E. W. Nester. 1981. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell 27:143-153. [DOI] [PubMed] [Google Scholar]
- 20.Gosseringer, R., E. Kuster, A. Galinier, J. Deutscher, and W. Hillen. 1997. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J. Mol. Biol. 266:665-676. [DOI] [PubMed] [Google Scholar]
- 21.Han, D. C., C. Y. Chen, Y. F. Chen, and S. C. Winans. 1992. Altered-function mutations of the transcriptional regulatory gene virG of Agrobacterium tumefaciens. J. Bacteriol. 174:7040-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heath, J. D., T. C. Charles, and E. W. Nester. 1995. Ti plasmid and chromosomally encoded two-component systems important in plant cell transformation by Agrobacterium species, p. 367-386. In J. Hoch and T. J. A. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 23.Hsieh, M., P. Hensley, M. Brenowitz, and J. S. Fetrow. 1994. A molecular model of the inducer binding domain of the galactose repressor of Escherichia coli. J. Biol. Chem. 269:13825-13835. [PubMed] [Google Scholar]
- 24.Hyytiainen, H., S. Sjoblom, T. Palomaki, A. Tuikkala, and E. T. Palva. 2003. The PmrA-PmrB two-component system responding to acidic pH and iron controls virulence in the plant pathogen Erwinia carotovora ssp. carotovora. Mol. Microbiol. 50:795-807. [DOI] [PubMed] [Google Scholar]
- 25.Lee, K., M. W. Dudley, K. M. Hess, D. G. Lynn, R. D. Joerger, and A. N. Binns. 1992. Mechanism of activation of Agrobacterium virulence genes: identification of phenol-binding proteins. Proc. Natl. Acad. Sci. USA 89:8666-8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, L., Y. Jia, Q. Hou, T. C. Charles, E. W. Nester, and S. Q. Pan. 2002. A global pH sensor: Agrobacterium sensor protein ChvG regulates acid-inducible genes on its two chromosomes and Ti plasmid. Proc. Natl. Acad. Sci. USA 99:12369-12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohrke, S. M., H. Yang, and S. Jin. 2001. Reconstitution of acetosyringone-mediated Agrobacterium tumefaciens virulence gene expression in the heterologous host Escherichia coli. J. Bacteriol. 183:3704-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mantis, N. J., and S. C. Winans. 1992. The Agrobacterium tumefaciens vir gene transcriptional activator virG is transcriptionally induced by acid pH and other stress stimuli. J. Bacteriol. 174:1189-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLean, B. G., E. A. Greene, and P. C. Zambryski. 1994. Mutants of Agrobacterium VirA that activate vir gene expression in the absence of the inducer acetosyringone. J. Biol. Chem. 269:2645-2651. [PubMed] [Google Scholar]
- 30.Melchers, L. S., T. J. Regensburg-Tuink, R. B. Bourret, N. J. Sedee, R. A. Schilperoort, and P. J. Hooykaas. 1989. Membrane topology and functional analysis of the sensory protein VirA of Agrobacterium tumefaciens. EMBO J. 8:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Mukhopadhyay, A., R. Gao, and D. G. Lynn. 2004. Integrating input from multiple signals: the VirA/VirG two-component system of Agrobacterium tumefaciens. Chembiochem 5:1535-1542. [DOI] [PubMed] [Google Scholar]
- 33.Palmer, A. G., R. Gao, J. Maresh, W. K. Erbil, and D. G. Lynn. 2004. Chemical biology of multi-host/pathogen interactions: chemical perception and metabolic complementation. Annu. Rev. Phytopathol. 42:439-464. [DOI] [PubMed] [Google Scholar]
- 34.Park, Y., Y. J. Cho, T. Ahn, and C. Park. 1999. Molecular interactions in ribose transport: the binding protein module symmetrically associates with the homodimeric membrane transporter. EMBO J. 18:4149-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pazour, G. J., C. N. Ta, and A. Das. 1992. Constitutive mutations of Agrobacterium tumefaciens transcriptional activator virG. J. Bacteriol. 174:4169-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng, W. T., Y. W. Lee, and E. W. Nester. 1998. The phenolic recognition profiles of the Agrobacterium tumefaciens VirA protein are broadened by a high level of the sugar binding protein ChvE. J. Bacteriol. 180:5632-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimoda, N., A. Toyoda-Yamamoto, S. Aoki, and Y. Machida. 1993. Genetic evidence for an interaction between the VirA sensor protein and the ChvE sugar-binding protein of Agrobacterium. J. Biol. Chem. 268:26552-26558. [PubMed] [Google Scholar]
- 38.Shimoda, N., A. Toyoda-Yamamoto, J. Nagamine, S. Usami, M. Katayama, Y. Sakagami, and Y. Machida. 1990. Control of expression of Agrobacterium vir genes by synergistic actions of phenolic signal molecules and monosaccharides. Proc. Natl. Acad. Sci. USA 87:6684-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stachel, S. E., G. An, C. Flores, and E. W. Nester. 1985. A Tn3 lacZ transposon for the random generation of β-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 4:891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stachel, S. E., E. Messens, M. Van Montagu, and P. Zambryski. 1986. Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature 318:624-629. [Google Scholar]
- 41.Toyoda-Yamamoto, A., N. Shimoda, and Y. Machida. 2000. Genetic analysis of the signal-sensing region of the histidine protein kinase VirA of Agrobacterium tumefaciens. Mol. Gen. Genet. 263:939-947. [DOI] [PubMed] [Google Scholar]
- 42.Tzfira, T., and V. Citovsky. 2002. Partners-in-infection: host proteins involved in the transformation of plant cells by Agrobacterium. Trends Cell Biol. 12:121-129. [DOI] [PubMed] [Google Scholar]
- 43.Umemura, T., Y. Matsumoto, K. Ohnishi, M. Homma, and I. Kawagishi. 2002. Sensing of cytoplasmic pH by bacterial chemoreceptors involves the linker region that connects the membrane-spanning and the signal-modulating helices. J. Biol. Chem. 277:1593-1598. [DOI] [PubMed] [Google Scholar]
- 44.Vyas, N. K., M. N. Vyas, and F. A. Quiocho. 1988. Sugar and signal-transducer binding sites of the Escherichia coli galactose chemoreceptor protein. Science 242:1290-1295. [DOI] [PubMed] [Google Scholar]
- 45.Wang, Y. 1999. Mechanism of signal integration and transmission mediating virulence induction in A. tumefaciens. Ph.D. thesis. University of Chicago, Chicago, Ill.
- 46.Wang, Y., R. Gao, and D. G. Lynn. 2002. Ratcheting up vir gene expression in Agrobacterium tumefaciens: coiled coils in histidine kinase signal transduction. Chembiochem 3:311-317. [DOI] [PubMed] [Google Scholar]
- 47.Wang, Y., A. Mukhopadhyay, V. R. Howitz, A. N. Binns, and D. G. Lynn. 2000. Construction of an efficient expression system for Agrobacterium tumefaciens based on the coliphage T5 promoter. Gene 242:105-114. [DOI] [PubMed] [Google Scholar]
- 48.Watson, B., T. C. Currier, M. P. Gordon, M. D. Chilton, and E. W. Nester. 1975. Plasmid required for virulence of Agrobacterium tumefaciens. J. Bacteriol. 123:255-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winans, S. C. 1990. Transcriptional induction of an Agrobacterium regulatory gene at tandem promoters by plant-released phenolic compounds, phosphate starvation, and acidic growth media. J. Bacteriol. 172:2433-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winans, S. C. 1992. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol. Rev. 56:12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winans, S. C., R. A. Kerstetter, and E. W. Nester. 1988. Transcriptional regulation of the virA and virG genes of Agrobacterium tumefaciens. J. Bacteriol. 170:4047-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winans, S. C., N. J. Mantis, C. Y. Chen, C. H. Chang, and D. C. Han. 1994. Host recognition by the VirA, VirG two-component regulatory proteins of Agrobacterium tumefaciens. Res. Microbiol. 145:461-473. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, J. 2000. At the Agrobacterium-maize interface: the chemical biology of pathogenesis. Ph.D. thesis. University of Chicago, Chicago, Ill.
- 54.Zhu, J., P. M. Oger, B. Schrammeijer, P. J. Hooykaas, S. K. Farrand, and S. C. Winans. 2000. The bases of crown gall tumorigenesis. J. Bacteriol. 182:3885-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]