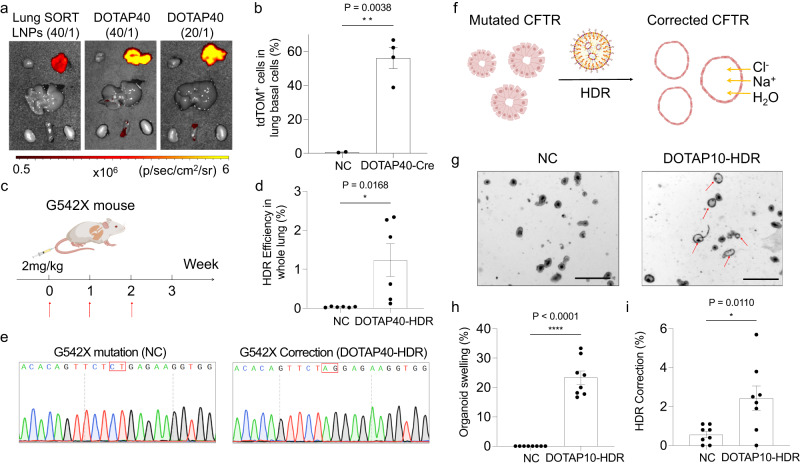
Fig. 4. DOTAP LNPs successfully corrected CFTR gene mutation in G542X-CF mouse model and restored CFTR function in a mouse intestinal organoid cell model with the G542X mutation.
a Optimized DOTAP40 LNPs showed higher luciferase expression in mouse lungs, compared to previously reported Lung SORT LNPs, and mRNA delivery efficiency remained comparable when the weight ratio of total lipid/total NA was reduced from 40/1 to 20/1 (0.1 mg kg−1 Luc mRNA, n = 3 biologically independent animals). b DOTAP40 LNPs efficiently delivered Cre mRNA (2 mg kg−1 Cre mRNA, 20/1) to basal cells of tdTOM mouse lung, activating tdTOM fluorescence. Two days after the second injection, the tdTOM positive cell in the mouse lung basal cell (NGFR + ) population was analyzed using flow cytometry (n = 4 biologically independent animals in DOTAP40-Cre group). NP only group was used as negative control (NC). c DOTAP40-HDR LNPs encapsulating Cas9 mRNA, sgRNA, and HDR template were I.V. injected into CF mouse containing G542X mutation over three weeks (2 mg kg−1 total NA, 20:1). d DOTAP40-HDR treatment successfully corrected G542X mutation in mouse lungs analyzed by NGS deep sequencing and CRISPResso2 analysis. NP only group was used as negative control (NC). Data are shown as mean±s.e.m. (n = 6 biologically independent animals). e Representative sequencing result showed the successful HDR correction event at G542X mutation site was observed in mouse lungs after DOTAP40-HDR, by subsequent cloning and DNA sequencing. f Schematic illustration shows the mechanism of ex vivo intestinal organoid based forksolin-induced swelling (FIS) assay. g Intestinal organoid swelling was observed after treatment of DOTAP10-HDR, suggesting successful CFTR function restoration. No organoid swelling was detected after NC (NP only) treatment. Scale bar: 500 μm. The data was repeated three times independently with similar results. More than 20% of organoids swelled (h) and successful HDR correction by Sanger sequencing (i) were detected after treated with DOTAP10-HDR. Data are shown as mean ± s.e.m. (n = 8 biologically independent samples). Two-tailed unpaired t-tests were used to determine the significance of the comparisons of data (*P < 0.05; **P < 0.01; ***P < 0.001). Source data are provided as a Source Data File.