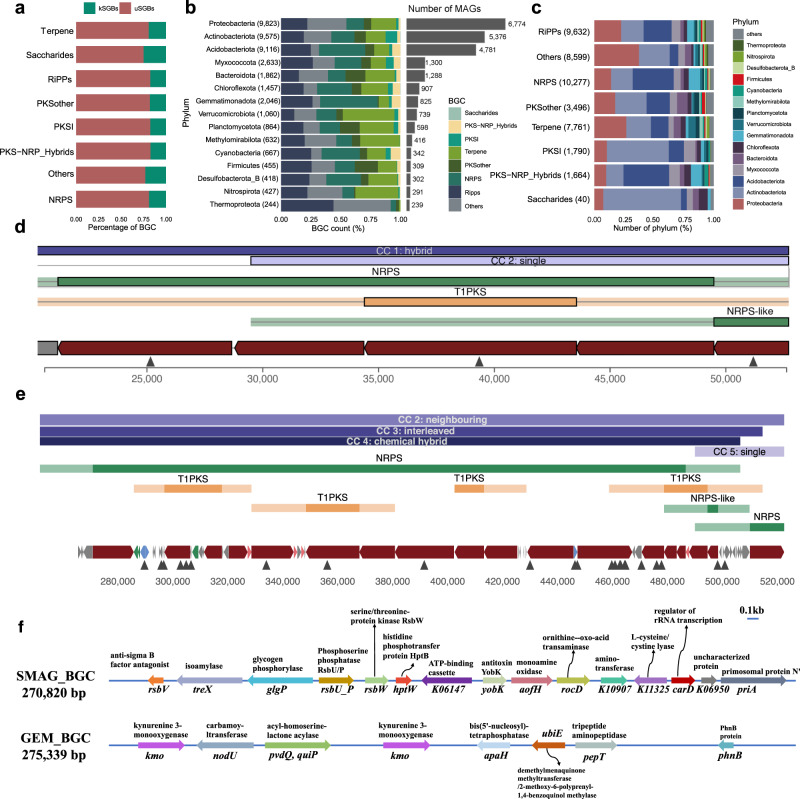
Fig. 4. Biosynthetic gene clusters recovered from the SMAG catalogue.
a BGCs of the SMAG between kSGBs and uSGBs. All the BGCs were separated into eight BiG-SCAPE classes. Non-ribosomal peptide synthetase (NRPS), Ribosomally synthesized and post-translationally modified peptide (RiPPs), polyketide synthase (PKS I), Terpene, PKS–NRPS hybrid, PKS other, Saccharides, Others. b The relative frequency of BGC types across dominant phyla BGC genes are predominantly identified in Proteobacteria, Actinobacteriota, Acidobacteriota and Bacteroidota. They are highly variable across phyla. c Number and BGC types identified from the SMAG. d Encoding the most remarkable number of BGC clusters including 111 NRPS or PKS modules and with clear colinear module chains. e The single largest BGC region found in a soil-derived bacterium from the Acidobacteria phylum and UBA5704 family. f Distribution and KO assignment of the two largest BGCs from SMAG and GEM.